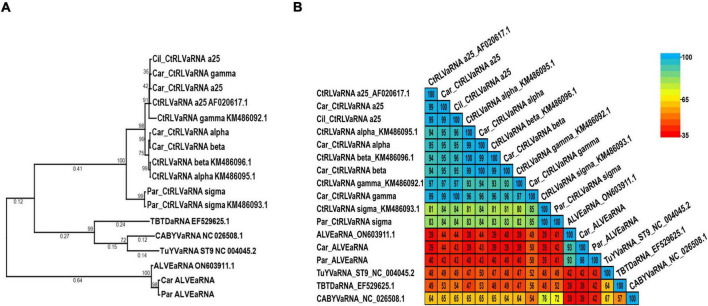
FIGURE 7.
Phylogenetic trees and average percent pairwise matrices depicting relationships of the tlaRNA sequences found in this study. Depicted are a maximum likelihood phylogenetic tree (A) and percent pairwise identity matrix (B) constructed using the P1a+P1b RdRp translated amino acid sequences of the tombusvirus-like associated RNAs (tlaRNAs) identified in this study. Numbers below the tree branches indicate the amino acid substitutions per site, and numbers above the lines indicate the bootstrap support values; bootstrap values below 50% are not shown (trees were calculated using 1000 bootstrap replicates). No species demarcation criteria have been established for tlaRNAs. References sequences are indicated by their GenBank accession numbers to the right of the virus label; accession numbers for the viruses identified in this study are listed in Table 4. CtRLVaRNA, carrot red leaf virus associated RNA. ALVEaRNA, arracacha latent virus E associated RNA. Outgroup sequences, TuYVaRNA ST9, turnip yellows virus aRNA ST9 TBTDaRNA, tobacco busy top disease aRNA; CABYVaRNA, cucurbit aphid borne yellows virus aRNA.