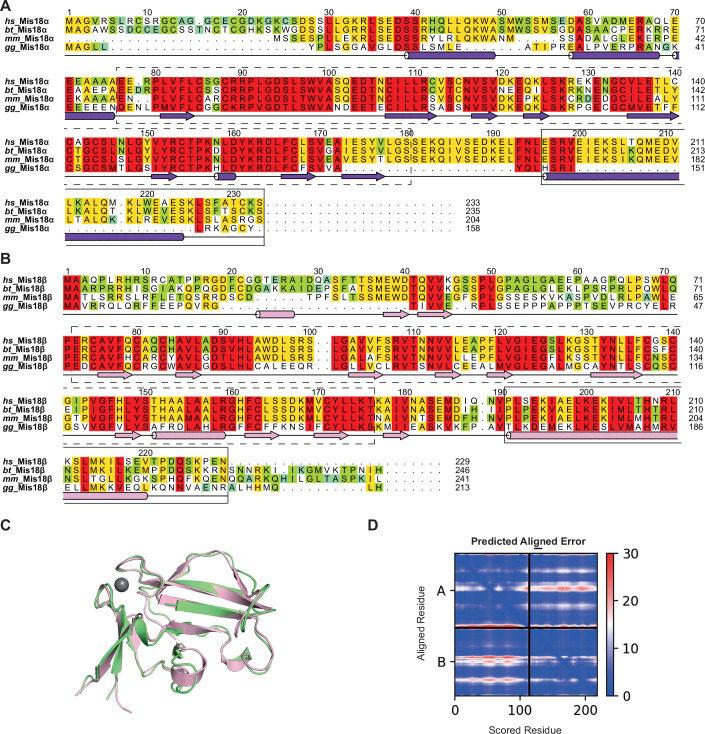
Figure EV1. Mis18α and Mis18β contain two domains capable of oligomerising.
(A, B) Domain architecture and amino acid conservation of (A) Mis18α and (B) Mis18β. Alignments include Homo sapiens (hs), Bos taurus (bt), Mus musculus (mm) and Gallus gallus (gg). The conservation score is mapped from red to cyan, where red corresponds to highly conserved and cyan to poorly conserved. Secondary structures as annotated/predicted by Conserved Domain Database [CDD] and PsiPred, http://bioinf.cs.ucl.ac.uk/psipred. Multiple sequence alignments were performed with MUSCLE (Madeira et al, 2019) and edited with Aline (Bond and Schüttelkopf, 2009). Dashed boxes highlight Yippee domains whilst solid boxes highlight C-terminus α-helices. (C) Superposition of Mis18βYippee structures predicted by AlphaFold (light pink) and RaptorX (green). RaptorX generated five models and the model with the lowest estimated error (1.9 Å) is shown here. The AlphaFold and RaptorX models superpose well with an RMSD of 0.95 Å. (D) The PAE plot corresponding to the Mis18α/βYippee AlphaFold model shown in Fig. 1D.