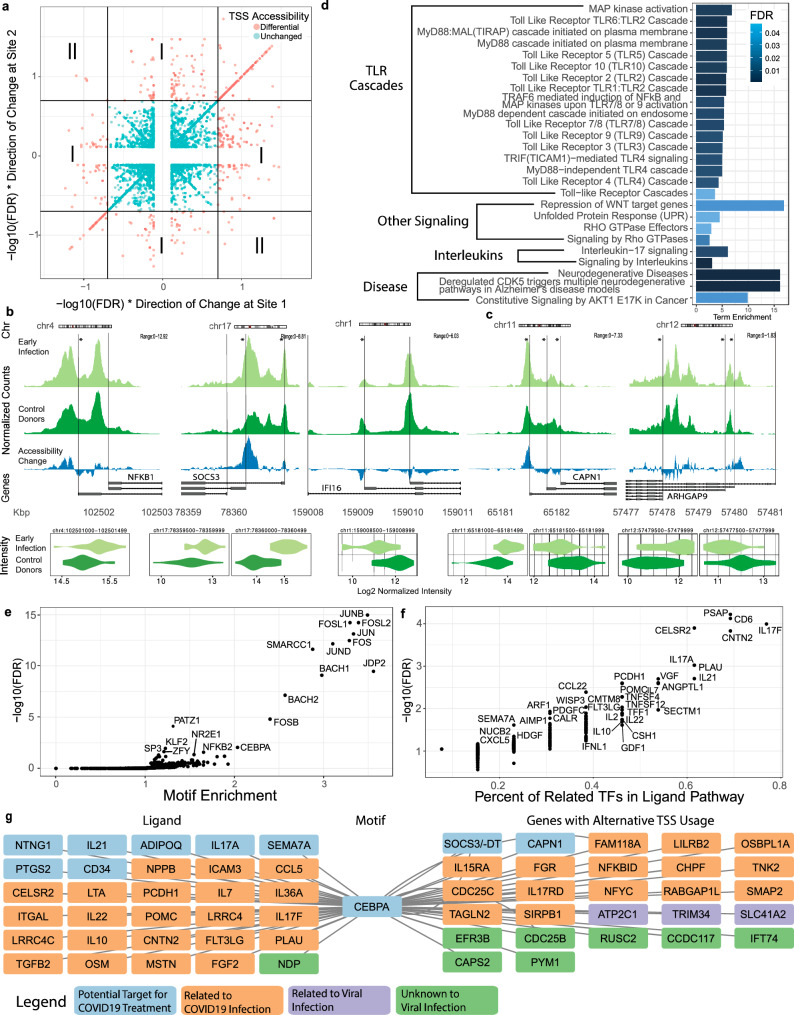
Fig. 4. Regulatory network construction on alternative transcription starting sites in CD16 monocytes during early COVID-19 infection.
a Scatter plot of differential accessibility at potential alternative transcription starting sites (TSSs). False discovery rate (FDR) and fold change (FC) were evaluated on chromatin accessibility in CD16 monocytes between COVID+ samples during early infection (n = 17) and COVID- samples (n = 22) in the COVID19X dataset. All pairwise combinations of -log10(FDR)*sign(log2(FC)) are shown. TSS pairs were categorized as type I if only one TSS was significantly differential (FDR < 0.2), or type II if both were significantly differential but in the opposite directions. Pairs of TSSs that were significantly differential in the same direction were not considered. b, c Coverage tracks illustrating type I (b) or type II (c) alternative TSS regulation around exemplar genes (* denotes a significant differential accessibility tile (DAT), FDR < 0.2). Precise FDR values are in the source data file. d Reactome pathway enrichment for genes with alternatively regulated TSSs (both type I and II). Pathway annotation was based on Reactome’s hierarchical database. e Motif enrichment using DATs involved in alternatively regulated TSSs and their co-accessible tiles (within ±1 M bp, zero-inflated correlation >0.5). f NicheNet-based ligand-motif set enrichment analysis (LMSEA) on motifs with FDR < 0.01. g A network centered around CEBPA that was constructed using significant ligands, motifs, and genes with alternatively regulated TSS sites. Ligand-motif links represent NicheNet-based associations. Motif-gene links represent motif presence in either an alternative TSS tile, or tiles correlated to an alternative TSS. TF transcription factor; Source data is provided in “SourceData_Figure4.xlsx”. Panels were generated using Adobe Illustrator.