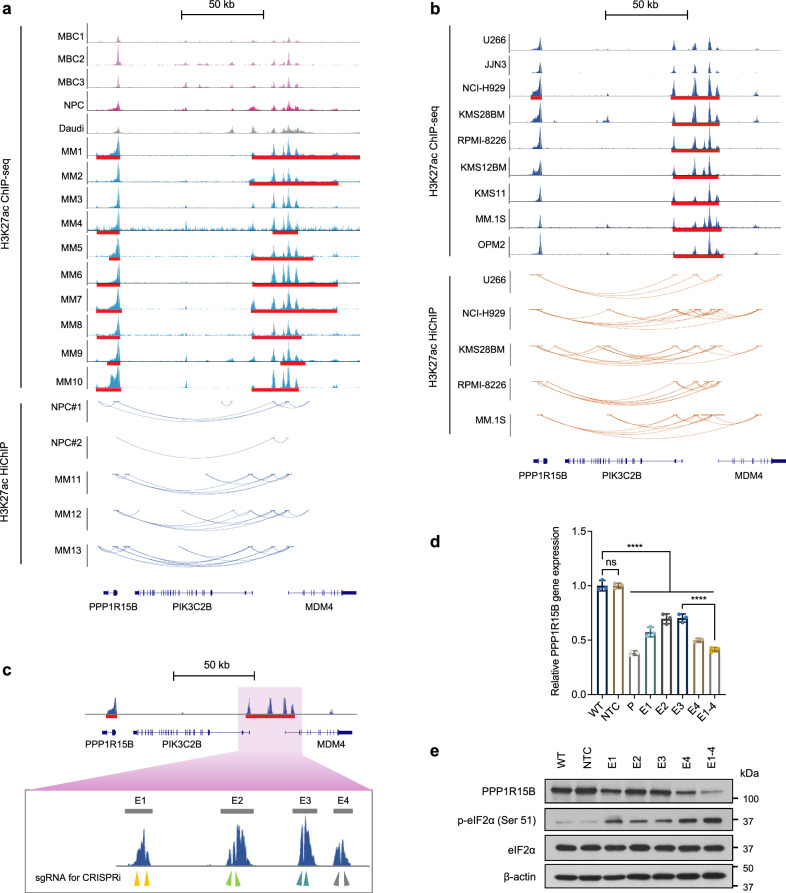
Fig. 4. PPP1R15B is a multiple myeloma-specific super-enhancer-associated gene.
a ChIP-seq around PPP1R15B gene and SE locus in patient-derived myeloma cells (MM1-10), and controls including three MBCs, one NPC sample and a Burkitt’s lymphoma cell line Daudi (top panel). SE broad peaks are marked by red lines. Enhancer-promoter linkages as determined by H3K27ac HiChIP are shown in patient-derived myeloma cells (n = 3) and NPCs from different donors (n = 2, bottom panel). b H3K27ac ChIP-seq signals at the PPP1R15B gene and SE locus in HMCLs (n = 9), with H3K27ac HiChIP interactions in HMCLs (n = 5) displayed below. c dCas9-KRAB sgRNA target sites at the constituent enhancers (E1-4) within the SE, as indicated by arrows. d, e RT-qPCR (d) and western blot analysis (e) of PPP1R15B expression in KMS28BM upon dCas9-KRAB-mediated repression of SE using sgRNAs targeting PPP1R15B promoter (P), individual SE constituents or in combination (E1-4). For RT-qPCR, one-way ANOVA with Tukey’s post hoc test was performed (p values in WT vs NTC = 1.00; WT vs P = 6.40 × 10-12; WT vs E1 = 1.55 × 10-9; WT vs E2 = 2.08 × 10-7; WT vs E3 = 2.83 × 10-7; WT vs E4 = 1.18 × 10-10; WT vs E1-4 = 1.32 × 10-11; E3 vs E1-4 = 4.05 × 10-7). n = 3 biological replicates. β-actin was used as a loading control. All western blots are representative of 3 biological replicates. Data are presented as mean ± SD. NPC, normal plasma cell; MBC memory B cell, NTC non-targeting control. Source data are provided as a Source Data file.