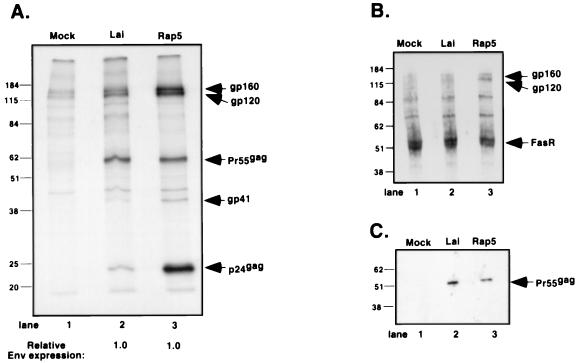
FIG. 5.
Effect of the vpu frameshift mutation on Env expression. (A) Jurkat cells (106) were infected with Lai or Rap at an MOI of 0.03. At 72 h postinfection, the cells were harvested and metabolically labeled with [35S]methionine-[35S]cysteine for 4 h. Viral proteins were immunoprecipitated with monoclonal antibodies against HIV-1 Env glycoproteins and HIV-1 Gag polyprotein. The amounts of gp160, gp120, gp41, p55gag, and p24gag were measured by PhosphorImage analysis, and the ratios of Env and Gag proteins relative to the Lai sample (lane 2) are indicated at the bottom of the fluorograph. (B) Jurkat cells (106) infected with Lai or Rap at an MOI of 0.03 were processed for cell surface biotinylation 72 h postinfection. Cultures (107 cells) were normalized for equal number of p24gag+ cells prior to biotinylation. Biotinylated extracts were immunoprecipitated with monoclonal antibodies against HIV-1 Env glycoproteins and cellular Fas receptor, electrophoresed on an SDS–10% polyacrylamide gel, blotted, and developed by ECL after incubation with streptavidin-horseradish peroxidase. (C) Western blot of extracts derived from Jurkat cells infected and processed as described for panel B. Solubilized proteins were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted with a monoclonal antibody against HIV-1 Gag polyprotein. Loading of samples was normalized according to the protein content of the extract. The positions of gp160, gp120, gp41, p55gag, p24gag, and Fas receptor are indicated on the right, and the positions of the molecular mass markers in kilodaltons are indicated on the left.