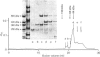Abstract
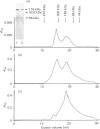
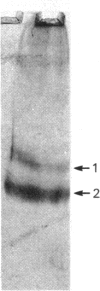
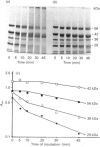
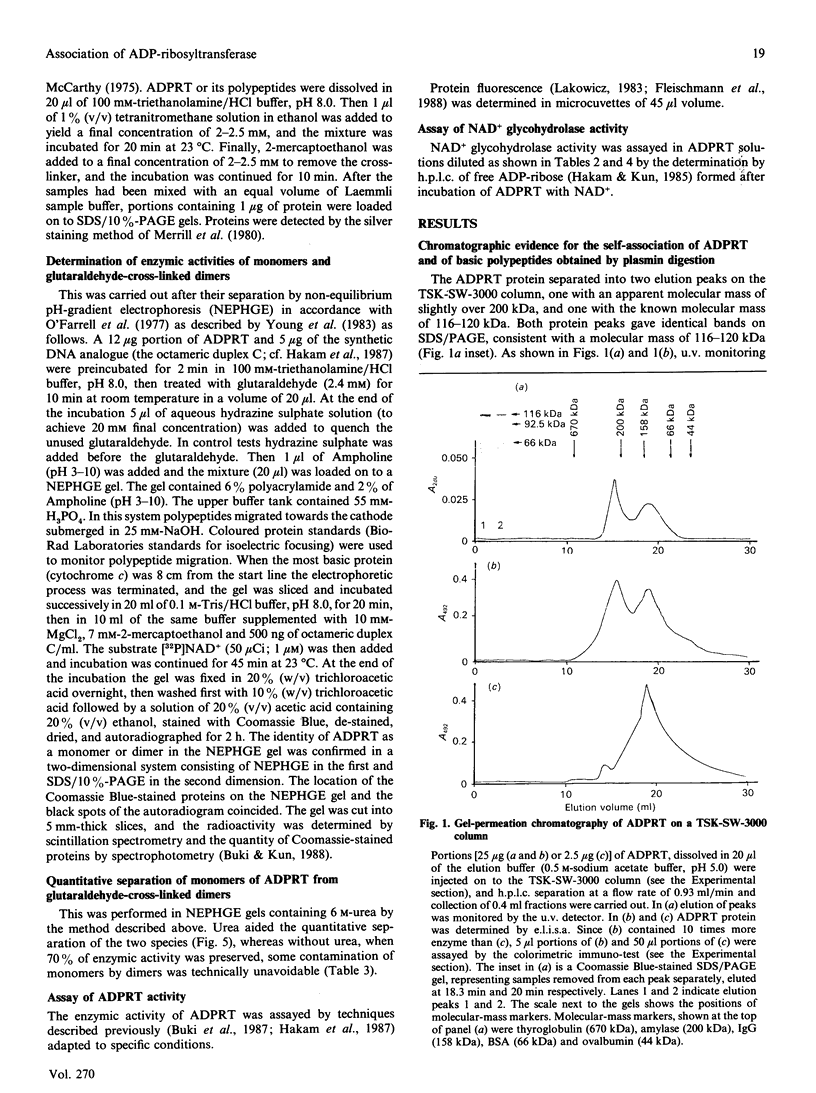
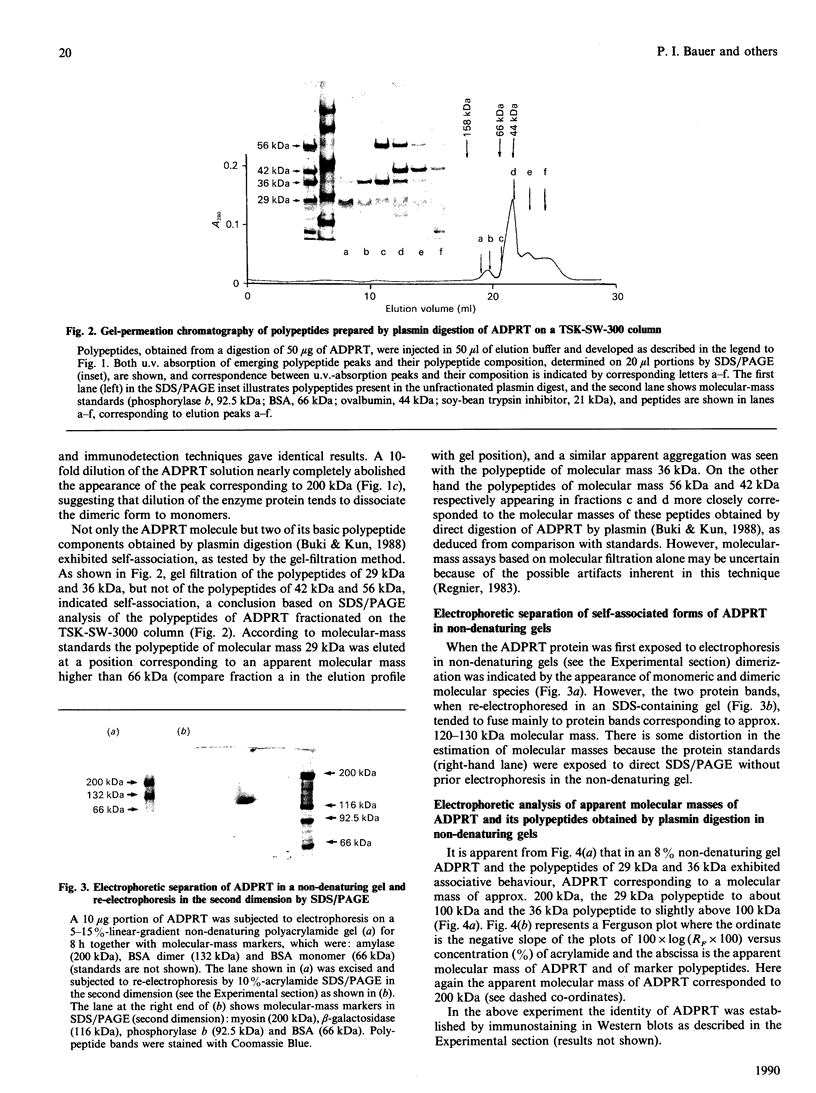
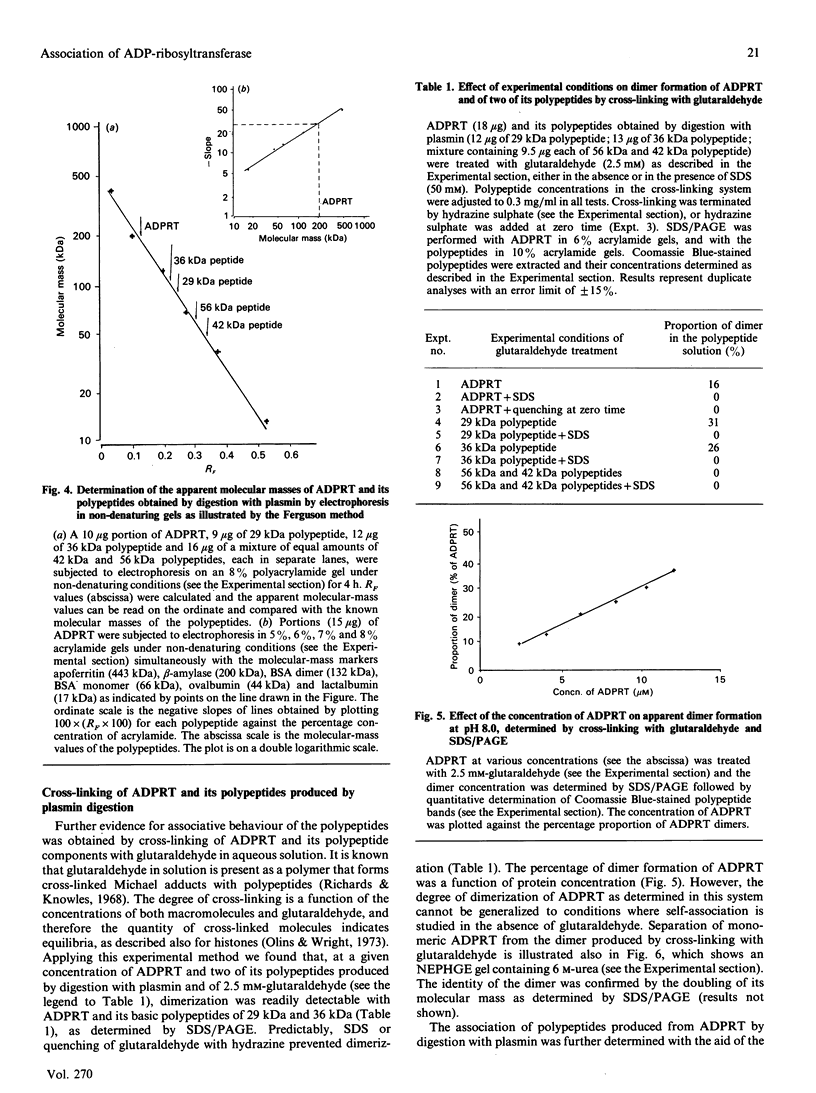
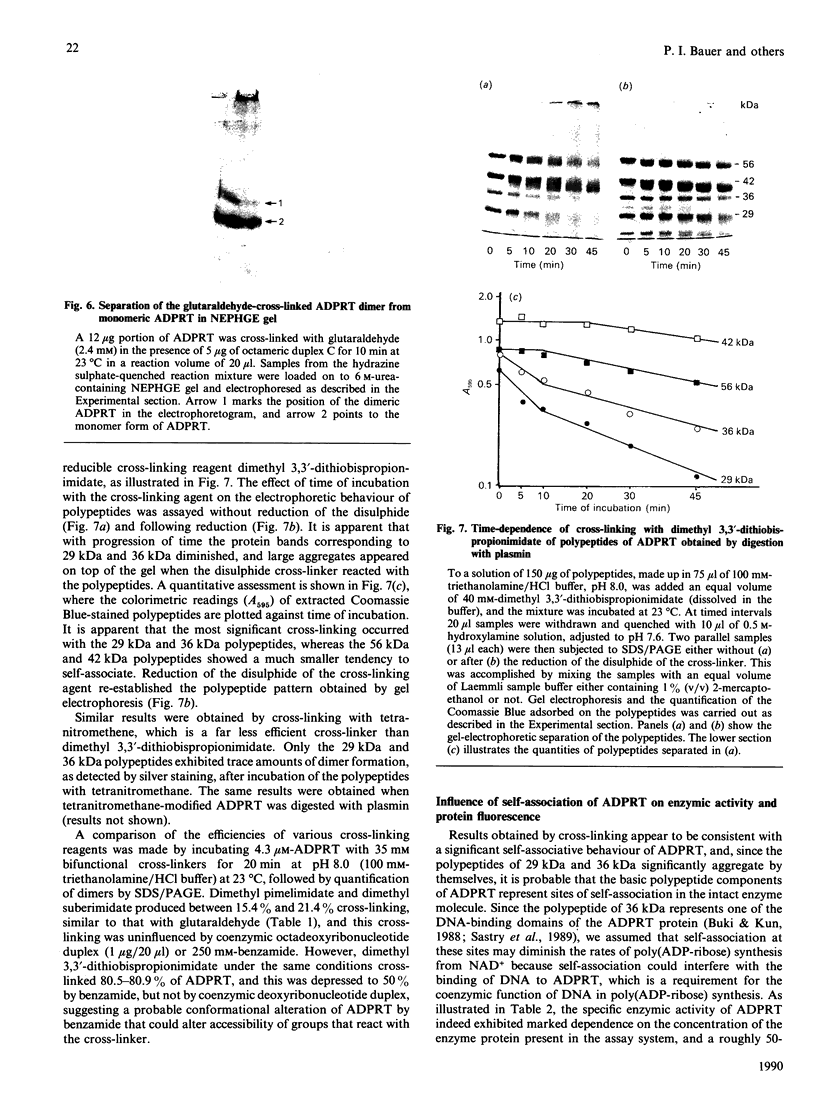
The macromolecular self-association of ADP-ribosyltransferase protein in solution was studied by several experimental techniques: quantitative gel filtration, electrophoretic analyses in non-denaturing gels, and cross-linking the enzyme protein with glutaraldehyde, dimethyl pimelimidate, dimethyl suberimidate, dimethyl 3,3'-dithiobisproprionimidate and tetranitromethane. The self-association of the polypeptide components obtained by plasmin digestion was also determined by using the above cross-linking agents. Monomers and cross-linked dimers of the enzyme protein, possessing enzymic activity, were separated in non-denaturing gels by electrophoresis. The basic polypeptide fragments, exhibiting molecular masses of 29 kDa and 36 kDa, self-associated, whereas the polypeptides with molecular masses of 56 kDa and 42 kDa associated only to a negligible extent, indicating that the peptide regions that also bind DNA and histones are probable sites of self-association in the intact enzyme molecule. Macromolecular association of the enzyme was indicated by a protein-concentration-dependent red-shift in protein fluorescence. The specific enzymic activity of the isolated ADP-ribosyltransferase depended on the concentration of the enzyme protein, and at 2.00 microM concentration the enzyme was self-inhibitory. Dilution of the enzyme protein to 30-40 nM resulted in a large increase in its specific activity. Further dilution to 1-3 nM coincided with a marked decrease of specific activity. Direct enzymic assays of electrophoretically separated monomers and cross-linked dimers demonstrated that the dimer appears to be the active molecular species that catalyses poly(ADP-ribose) synthesis. The NAD+ glycohydrolase activity of the enzyme was also dependent on protein concentration and was highest at 1-3 nM enzyme concentration, when polymerase activity was minimal, indicating that the monomeric enzyme behaved as a glycohydrolase, whereas poly(ADP-ribosyl)ation of enzyme molecules was maximal when the enzyme tends to be self-associated to the dimeric form.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-Ela N., Jacobson E. L., Jacobson M. K. Labeling methods for the study of poly- and mono(ADP-ribose) metabolism in cultured cells. Anal Biochem. 1988 Oct;174(1):239–250. doi: 10.1016/0003-2697(88)90541-6. [DOI] [PubMed] [Google Scholar]
- Bauer P. I., Hakam A., Kun E. Mechanisms of poly(ADP-ribose) polymerase catalysis; mono-ADP-ribosylation of poly(ADP-ribose) polymerase at nanomolar concentrations of NAD. FEBS Lett. 1986 Jan 20;195(1-2):331–338. doi: 10.1016/0014-5793(86)80188-0. [DOI] [PubMed] [Google Scholar]
- Buki K. G., Kirsten E., Kun E. Isolation of adenosine diphosphoribosyltransferase by precipitation with reactive red 120 combined with affinity chromatography. Anal Biochem. 1987 Nov 15;167(1):160–166. doi: 10.1016/0003-2697(87)90147-3. [DOI] [PubMed] [Google Scholar]
- Buki K. G., Kun E. Polypeptide domains of ADP-ribosyltransferase obtained by digestion with plasmin. Biochemistry. 1988 Aug 9;27(16):5990–5995. doi: 10.1021/bi00416a024. [DOI] [PubMed] [Google Scholar]
- Chrambach A., Rodbard D. Polyacrylamide gel electrophoresis. Science. 1971 Apr 30;172(3982):440–451. doi: 10.1126/science.172.3982.440. [DOI] [PubMed] [Google Scholar]
- FERGUSON K. A. STARCH-GEL ELECTROPHORESIS--APPLICATION TO THE CLASSIFICATION OF PITUITARY PROTEINS AND POLYPEPTIDES. Metabolism. 1964 Oct;13:SUPPL–SUPPL1002. doi: 10.1016/s0026-0495(64)80018-4. [DOI] [PubMed] [Google Scholar]
- Fleischmann J. D., Wentworth D., Valencic F., Imbembo A. L., Koehler K. A. Interleukin-2 self-association. Biochem Biophys Res Commun. 1988 Apr 29;152(2):879–885. doi: 10.1016/s0006-291x(88)80121-9. [DOI] [PubMed] [Google Scholar]
- Gaal J. C., Pearson C. K. Eukaryotic nuclear ADP-ribosylation reactions. Biochem J. 1985 Aug 15;230(1):1–18. doi: 10.1042/bj2300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakam A., Kun E. High-performance liquid chromatography of in vitro synthesized poly(ADP-ribose) on ion-exchange columns, separation of oligomers of varying chain length and estimation of apparent branching. J Chromatogr. 1985 Aug 23;330(2):287–298. doi: 10.1016/s0021-9673(01)81985-3. [DOI] [PubMed] [Google Scholar]
- Hakam A., McLick J., Buki K., Kun E. Catalytic activities of synthetic octadeoxyribonucleotides as coenzymes of poly(ADP-ribose) polymerase and the identification of a new enzyme inhibitory site. FEBS Lett. 1987 Feb 9;212(1):73–78. doi: 10.1016/0014-5793(87)81559-4. [DOI] [PubMed] [Google Scholar]
- Hermann R., Rubolph R., Jaenicke R. Kinetics of in vitro reconstitution of oligomeric enzymes by cross-linking. Nature. 1979 Jan 18;277(5693):243–245. doi: 10.1038/277243a0. [DOI] [PubMed] [Google Scholar]
- Honegger A. M., Kris R. M., Ullrich A., Schlessinger J. Evidence that autophosphorylation of solubilized receptors for epidermal growth factor is mediated by intermolecular cross-phosphorylation. Proc Natl Acad Sci U S A. 1989 Feb;86(3):925–929. doi: 10.1073/pnas.86.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikejima M., Marsischky G., Gill D. M. Direction of elongation of poly(ADP-ribose) chains. Addition of residues at the polymerase-proximal terminus. J Biol Chem. 1987 Dec 25;262(36):17641–17650. [PubMed] [Google Scholar]
- Janssen O. E., Hilz H. Differentiation of 3T3-L1 pre-adipocytes induced by inhibitors of poly(ADP-ribose) polymerase and by related noninhibitory acids. Eur J Biochem. 1989 Apr 1;180(3):595–602. doi: 10.1111/j.1432-1033.1989.tb14686.x. [DOI] [PubMed] [Google Scholar]
- Kameshita I., Matsuda M., Nishikimi M., Ushiro H., Shizuta Y. Reconstitution and poly(ADP-ribosyl)ation of proteolytically fragmented poly(ADP-ribose) synthetase. J Biol Chem. 1986 Mar 15;261(8):3863–3868. [PubMed] [Google Scholar]
- Kristensen T., Holtlund J. Poly(ADP-ribose) polymerase from Ehrlich ascites tumor cells. Properties of the purified polymerase. Eur J Biochem. 1978 Aug 1;88(2):495–501. doi: 10.1111/j.1432-1033.1978.tb12475.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ludwig A., Behnke B., Holtlund J., Hilz H. Immunoquantitation and size determination of intrinsic poly(ADP-ribose) polymerase from acid precipitates. An analysis of the in vivo status in mammalian species and in lower eukaryotes. J Biol Chem. 1988 May 25;263(15):6993–6999. [PubMed] [Google Scholar]
- Lutz H. U., Lomant A. J., McMillan P., Wehrli E. Rearrangements of integral membrane components during in vitro aging of sheep erythrocyte membranes. J Cell Biol. 1977 Aug;74(2):389–398. doi: 10.1083/jcb.74.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MELLORS R. C., KUPFER A., HOLLENDER A. Quantitative cytology and cytopathology. I. Measurement of the thickness, the volume, the hydrous mass, and the anhydrous mass of living cells by interference microscopy. Cancer. 1953 Mar;6(2):372–384. doi: 10.1002/1097-0142(195303)6:2<372::aid-cncr2820060222>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Mandel P., Okazaki H., Niedergang C. Poly(adenosine diphosphate ribose). Prog Nucleic Acid Res Mol Biol. 1982;27:1–51. doi: 10.1016/s0079-6603(08)60596-6. [DOI] [PubMed] [Google Scholar]
- Martinson H. G., McCarthy B. J. Histone-histone associations within chromatin. Cross-linking studies using tetranitromethane. Biochemistry. 1975 Mar 11;14(5):1073–1078. doi: 10.1021/bi00676a030. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. Z., Goodman H. M., O'Farrell P. H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977 Dec;12(4):1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Olins D. E., Wright E. B. Glutaraldehyde fixation of isolated eucaryotic nuclei. Evidence for histone-histone proximity. J Cell Biol. 1973 Nov;59(2 Pt 1):304–317. doi: 10.1083/jcb.59.2.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin P. W., Jacobson E. L., Benjamin R. C., Moss J., Jacobson M. K. Quantitative studies of inhibitors of ADP-ribosylation in vitro and in vivo. J Biol Chem. 1989 Mar 15;264(8):4312–4317. [PubMed] [Google Scholar]
- Regnier F. E. High-performance liquid chromatography of proteins. Methods Enzymol. 1983;91:137–190. doi: 10.1016/s0076-6879(83)91016-9. [DOI] [PubMed] [Google Scholar]
- Richards F. M., Knowles J. R. Glutaraldehyde as a protein cross-linkage reagent. J Mol Biol. 1968 Oct 14;37(1):231–233. doi: 10.1016/0022-2836(68)90086-7. [DOI] [PubMed] [Google Scholar]
- Sastry S. S., Buki K. G., Kun E. Binding of adenosine diphosphoribosyltransferase to the termini and internal regions of linear DNAs. Biochemistry. 1989 Jun 27;28(13):5670–5680. doi: 10.1021/bi00439a050. [DOI] [PubMed] [Google Scholar]
- Sastry S. S., Kun E. Molecular interactions between DNA, poly(ADP-ribose) polymerase, and histones. J Biol Chem. 1988 Jan 25;263(3):1505–1512. [PubMed] [Google Scholar]
- Sóoki-Tóth A., Bánfalvi G., Szöllösi J., Kirsten E., Staub M., Antoni F., Kun E. Cellular regulation of ADP-ribosylation of proteins. III. Selective augmentation of in vitro ADP-ribosylation of histone H3 in murine thymic cells after in vivo emetine treatment. Exp Cell Res. 1989 Sep;184(1):44–52. doi: 10.1016/0014-4827(89)90362-5. [DOI] [PubMed] [Google Scholar]
- Tseng A., Jr, Lee W. M., Jakobovits E. B., Kirsten E., Hakam A., McLick J., Buki K., Kun E. Prevention of tumorigenesis of oncogene-transformed rat fibroblasts with DNA site inhibitors of poly(ADP ribose) polymerase. Proc Natl Acad Sci U S A. 1987 Feb;84(4):1107–1111. doi: 10.1073/pnas.84.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K., Hayaishi O. ADP-ribosylation. Annu Rev Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- Ushiro H., Yokoyama Y., Shizuta Y. Purification and characterization of poly (ADP-ribose) synthetase from human placenta. J Biol Chem. 1987 Feb 15;262(5):2352–2357. [PubMed] [Google Scholar]
- Yamanaka H., Penning C. A., Willis E. H., Wasson D. B., Carson D. A. Characterization of human poly(ADP-ribose) polymerase with autoantibodies. J Biol Chem. 1988 Mar 15;263(8):3879–3883. [PubMed] [Google Scholar]
- Young D. A., Voris B. P., Maytin E. V., Colbert R. A. Very-high-resolution two-dimensional electrophoretic separation of proteins on giant gels. Methods Enzymol. 1983;91:190–214. doi: 10.1016/s0076-6879(83)91017-0. [DOI] [PubMed] [Google Scholar]