FIGURE 1.
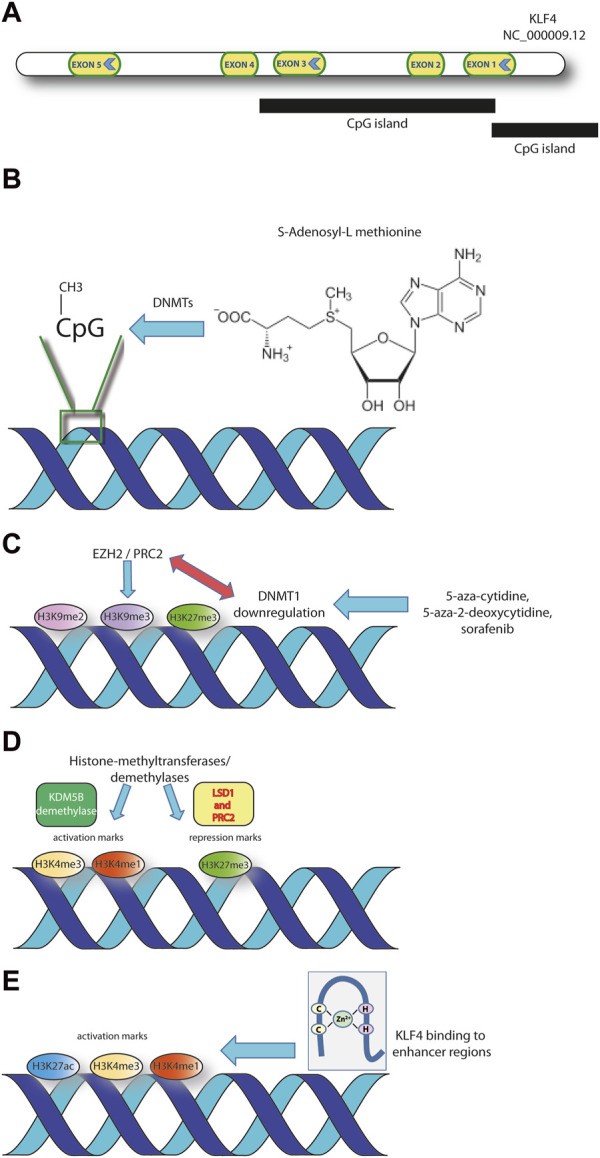
The main layers of KLF4 epigenetic regulation discussed in the text are represented. (A) Structure of KLF4 gene (GRCh38.p14 assembly). (B) Methylation at specific CpG sites within KLF4 CpG islands. S-adenosyl-methionine is the universal methyl group donor in DNA- and histone-methylation reactions. (C) Methylation of specific histone residues represent repressory marks at specific DNA sequences. The interaction of DNMT1 with histone methyltransferases affects the chromatin architecture. (D) Histone methylation on specific histone residues within regulatory regions may represent activatory or repressory marks. The process is finely tuned through histone methyltransferases and demethylases. (E) KLF4 binding to enhancer regions, enriched in histone activation marks (H3K27ac, H3K4me1, H3K4me3), is a characteristic of the regulatory footprint of some tumors.
