Fig. 2.
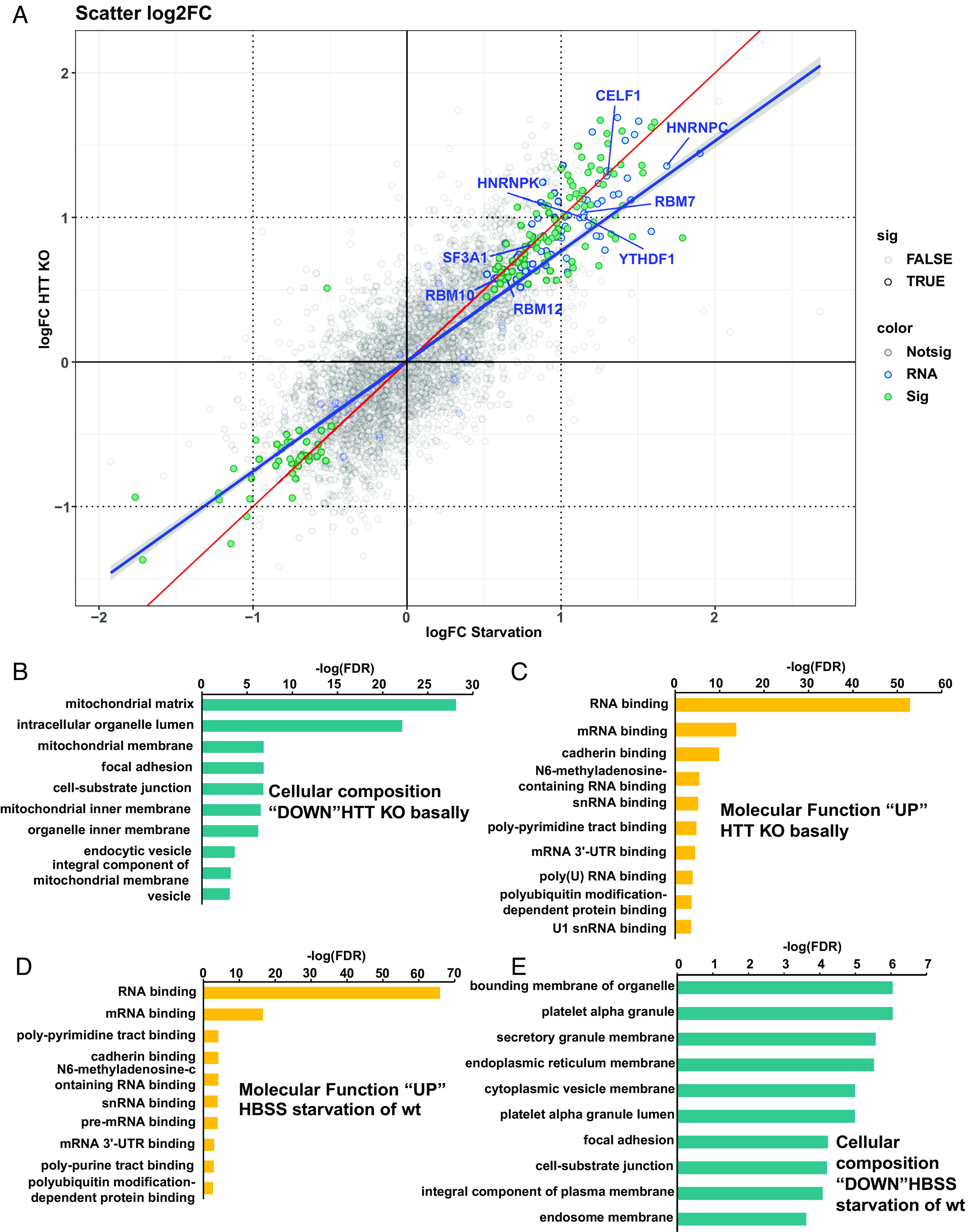
HTT KO alters the contents of lysosomes. LysoIP was performed in 8988T cells expressing TMEM192-HA using magnetic HA pulldown beads. All cells were treated with Baf for 4 h prior to harvest in order to preserve lysosomal proteins for analysis. Proteomic contents of purified lysosomes were analyzed by mass spectrometry. Scatterplot of log2 fold changes comparing HTT KO to WT starvation-induced macroautophagy (turquoise = significant in both lists, blue = RNA- or mRNA-binding protein, red line = perfect correlation, blue line = LOESS curve fit, labeled proteins are exemplary). (A). Gene ontology pathways significantly altered by HTT KO were identified using Enrichr. Threshold for significant enrichment was FDR < 0.1. (B) Cellular composition enrichment of proteins that were decreased in the lysosomes of HTT KO cells compared to parental cells. (C) Molecular function enrichment of proteins that were increased in the lysosomes of HTT KO cells compared to parental cells. (D) Molecular function enrichment of proteins that were increased in the lysosomes of parental cells with HBSS treatment for 4 h prior to harvest, for stimulation of macroautophagy. (E) Cellular composition enrichment of proteins that were decreased in the lysosomes of parental cells with HBSS treatment for 4 h prior to harvest did not include mitochondrial proteins, unlike what we observed with HTT KO basally.
