Fig. 3.
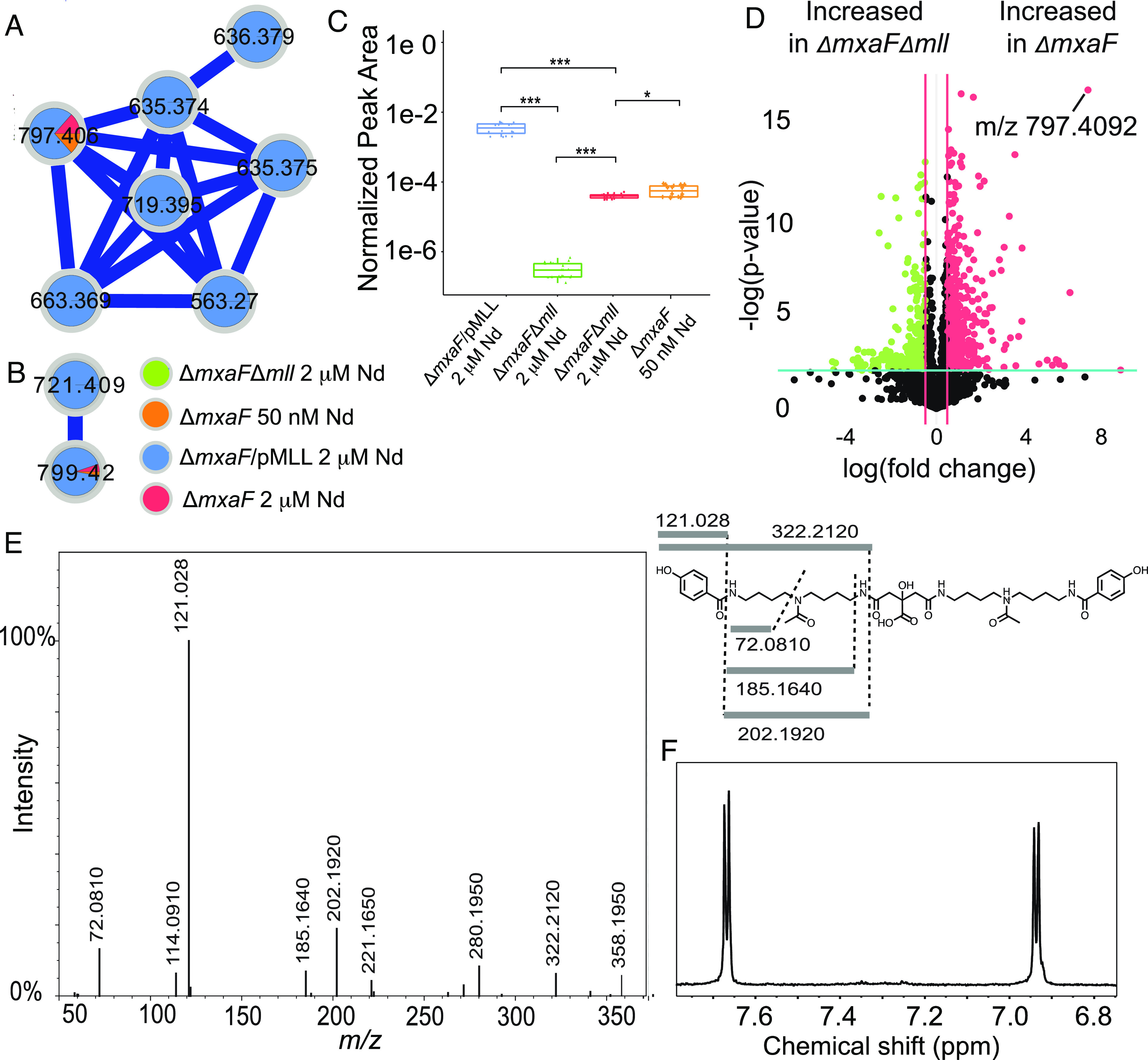
Identification of methylolanthanin via molecular networking and MS/MS. (A) ESI-UHPLC–MS/MS in negative ionization mode reveals a molecular family of MS/MS spectra only found in ΔmxaF (orange and red) and ΔmxaF/pMLL (blue) but not in ΔmxaFΔmll (green). (B) ESI-UHPLC-MS/MS in positive ionization mode reveals a molecular family of MS/MS spectra only found in ΔmxaF (orange and red) and ΔmxaF/pMLL (blue) but not in ΔmxaFΔmll (green). Two features (m/z 799.4232 in positive mode/m/z 797.4092 in negative mode and m/z 721.4114 in positive mode/m/z 719.3978 in negative mode) are found in both positive and negative ionization mode molecular families. (C) Boxplots of the normalized peak area of feature m/z 797.4092 in negative mode reveal significant differences between peak area in all conditions. Statistics were calculated with n = 20 per group Kruskal–Wallis, followed by pairwise Wilcoxon tests and Benjamini–Hochberg (BH) correction (**P < 0.001, *P < 0.05). Upper and lower whiskers extend to the closest value to ±1.5 * IQR. (D) m/z 797.4092 is one of the most significantly increased features in ΔmxaF vs. ΔmxaFΔmll supernatants when analyzed using ESI-UHPLC-MS/MS in positive ionization mode. Volcano plot analysis was performed with n = 5 per group. Fold changes 0.5 and a P-value < 0.01 are highlighted. (E) The consensus MS/MS spectrum in positive mode is consistent with the proposed structure, as highlighted by the key fragments in the chemical structure. (F) The NMR aromatic region of isolated methylolanthanin supports a parasubstituted monohydroxybenzoate moiety (1H NMR spectrum, 800 MHz, D2O + 0.003% TMSP).
