Fig. 5.
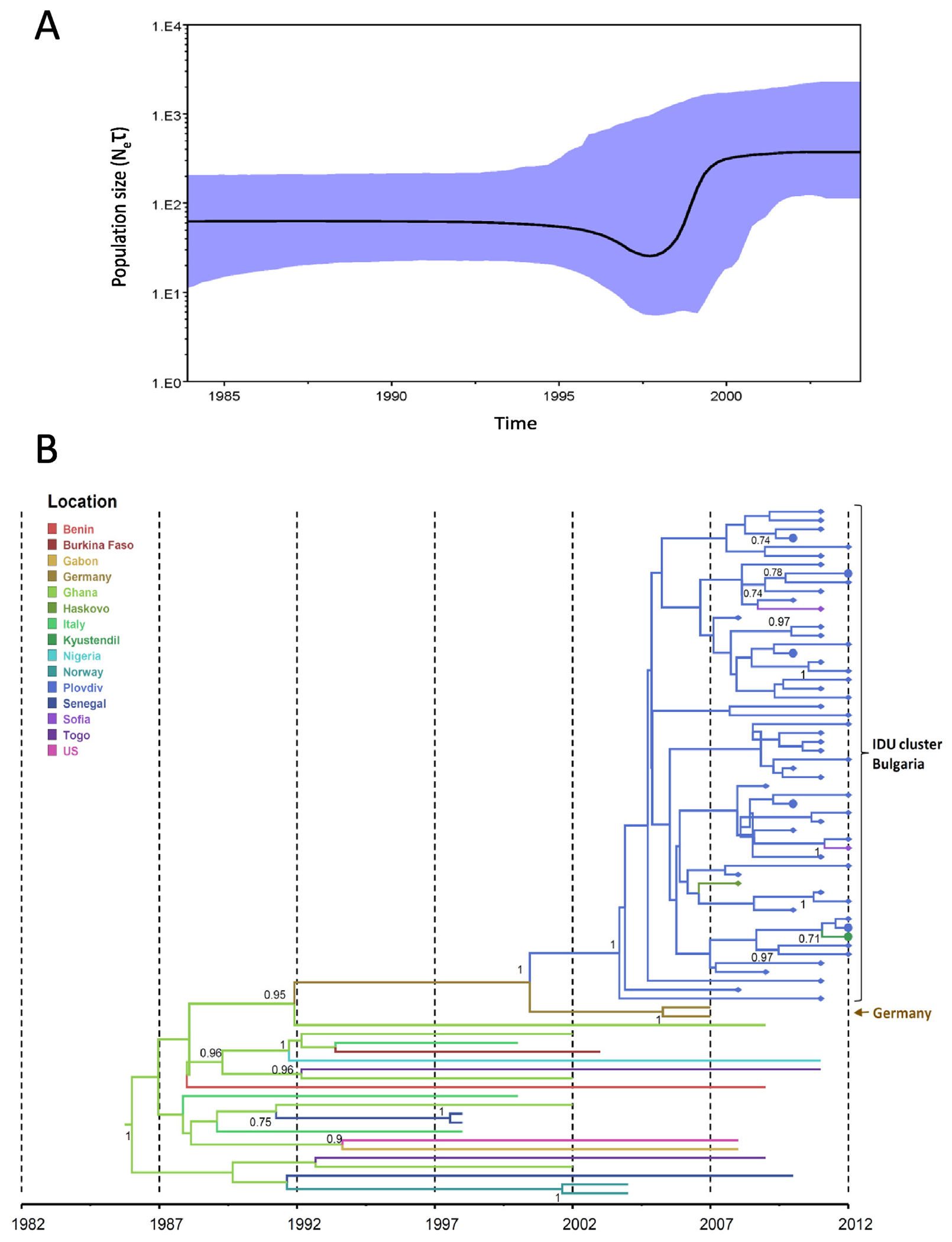
Phylodynamics and phylogeography of HIV-1 CRF02_AG in Bulgaria inferred by Bayesian analysis. (A) Bayesian skyride plot showing the inferred growth of the CRF02_AG epidemic in Bulgaria. Inferred effective population sizes (Ne) over time in years are on the y- and x-axes, respectively. (B) Phylogenetic tree showing the most recent common ancestor and geographic spread of CRF02_AG. Analysis included 69 polymerase (pol) sequences from Bulgaria, 50 sequences from persons who inject drugs (PWIDs), 19 heterosexual (HET) individuals and 22 pol sequences from other countries. Filled circles and diamonds at branch tips indicate Bulgarian HET and PWIDs, respectively. Time scale in years provided on x-axis and country or city origin of the sequences provided in color and defined in key.
