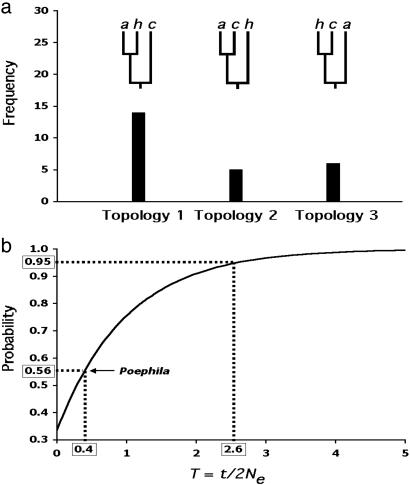
Fig. 1.
Conflicts between gene and species trees in Poephila finches, and their implications. (a) Frequency distribution of all three possible gene trees encountered in a survey of nuclear DNA sequence variation among three species of Australian grassfinches (Poephila). Branch tips are labeled as follows: a, P. acuticauda; h, P. hecki; c, P. cincta. P. cincta vs. P. acuticauda/hecki is a well established divergence across the Carpentarian barrier in northern Australia and is supported by numerous species-level phenotypic differences; P. hecki vs. acuticauda are allopatric but represent a more recent taxonomic split which is regarded by some (38) as phylogenetic species. In the genetic survey, a single allele was sampled from each of the three taxa for thirty presumably unlinked loci. Twenty-five gene trees exhibiting all three possible topologies were unambiguously reconstructed. Gene trees for five loci could not be resolved. Topology 1 reflects the presumed species tree. (b) Unconditional probabilities of a gene tree being congruent with the species tree over a range of values for T (after ref. 43). The curve is based on the equation for three species [P(congruent gene tree) = 1-2/3e-T (42, 43), where T is equal to t/2Ne, t is defined as the time between speciation events (in generations), and 2Ne is twice the size of the effective population size of the basal ancestor. When T = 0, the topology of a gene tree of three sampled alleles is expected to be random with respect to the species tree (i.e., probability of 0.33). Note that T must be at least as large as 2.6 for a 0.95 probability of congruence, yet the empirically derived probability for Poephila finches of 0.56 results in a T value of only 0.4. The Poephila tree congruence probability is based on the fact that 14 of 25 independent gene trees matched the presumed species tree observed by W.B.J. and S.V.E. (unpublished work).