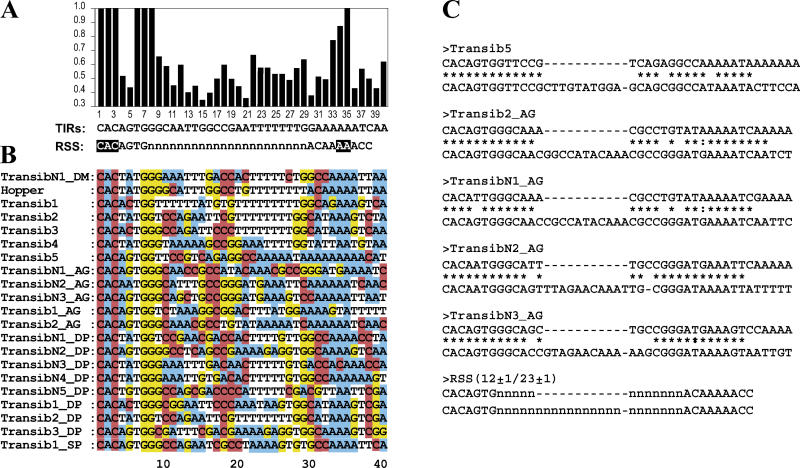
Figure 4. Structural Similarities between the Transib TIRs and V(D)J RSS Signals.
The species abbreviations are: AA, yellow fever mosquito; AG, African malaria mosquito; DM, D. melanogaster fruit fly DP, D. pseudoobscura fruit fly; SP, sea urchin. (Transib1 through Transib5 are from the fruit fly D. melanogaster).
(A) Frequencies of the most frequent nucleotides at each position of the consensus sequence of the 5′ TIRs of transposons that belong to 20 families of Transib transposons identified in fruit flies and mosquitoes. The RSS23 consensus sequence is shown immediately under the TIRs consensus sequence. The most conserved nucleotides in the RSS23 heptamer and nonamer, which are necessary for efficient V(D)J recombination, are highlighted. The 23 ± 1 bp variable spacer is marked by Ns.
(B) Non-gapped alignment of consensus sequences of 5′ TIRs from 21 families of Transib transposons.
(C) The 12/23 rule follows from the basic structure of TIRs of the consensus sequences of transposons that belong to the Transib5, Transib2_AG, TransibN1_AG, TransibN2_AG, and TransibN3_AG families. The 5′ TIRs of these transposons are aligned with the corresponding 3′ TIRs. Structures of the 5′ and 3′ TIRs resemble RSS12 and RSS23, respectively.