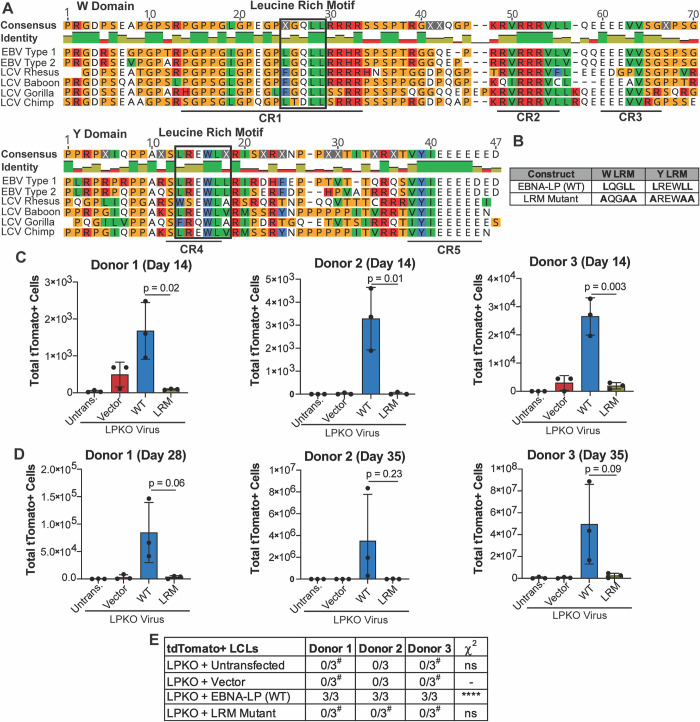
Fig 3. EBNA-LP contains Leucine-rich motifs which are important for adult naïve B cell transformation.
A. Alignment of EBNA-LP W and Y domains across human EBV (Type 1 is B95-8, Type 2 is AG876) and LCV strains, with identified leucine-rich motifs highlighted in black boxes. Identity bar indicates conservation of amino acids, with green indicating highly conserved regions, yellow indicating less conserved, and red are poorly conserved regions. Colors of residues indicate amino acid properties. Previously defined conserved regions (CR1, CR2, CR3, CR4, CR5) are underlined (22). B. LRM Mutant construct generated with mutations in the leucine-rich domains has modified leucines to alanines. C. Total tdTomato positive cells at 14 days post infection for each donor in LPKO virus infected, trans-complemented cells (n = 3). D. Total tdTomato positive cells in adult naive B cells from three donors at 28 or 35 days post infection (n = 3). P values from unpaired t-test. Mean and standard deviation are plotted. E. Total tdTomato positive LCLs generated from trans-complemented LPKO infected adult naïve B cells. #Indicates conditions in which some replicates generated tdTomato negative LCLs. P values calculated using Fisher’s exact test to compare transformation outcomes to LPKO + Vector. **** indicates a p value < 0.0001.