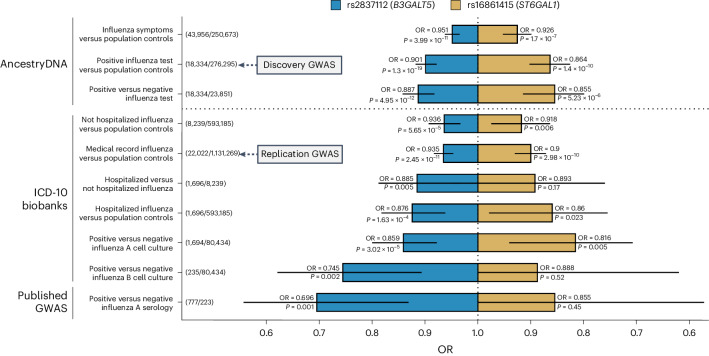
Fig. 2. Association between ST6GAL1 and B3GALT5 variants and ten influenza-related phenotypes.
Variants in ST6GAL1 (rs16861415) and B3GALT5 (rs2837112) were associated with lower risk of reporting a positive test for influenza in the AncestryDNA cohort (discovery GWAS, total n = 294,629). The association between both variants and influenza infection was confirmed when analyzing medical record-based influenza status in an independent analysis of 1,153,291 individuals from seven biobank cohorts (replication GWAS; the cohorts are listed in Supplementary Table 1). Sensitivity analyses based on eight additional phenotypes showed that (1) in the AncestryDNA cohort, effect sizes for both variants were comparable after excluding controls with no available influenza test results, while they were weaker when testing a looser phenotype that considered only flu-like symptoms; (2) in two biobank cohorts with available hospitalization data (UKB, GHS), restricting the case group to individuals with influenza-related hospitalization resulted in stronger effect sizes for both variants, with the B3GALT5 variant significantly reducing the risk of hospitalization among infected cases; and (3) consistent and often stronger (by effect size) associations were observed with phenotypes that captured recent influenza infection, such as a positive cell culture or serology test for influenza A. Further details on these associations are provided in Supplementary Table 6. Unadjusted P values were derived using Firth regression (two-sided test) implemented in REGENIE9. The error bars represent the 95% CI for the OR estimate.