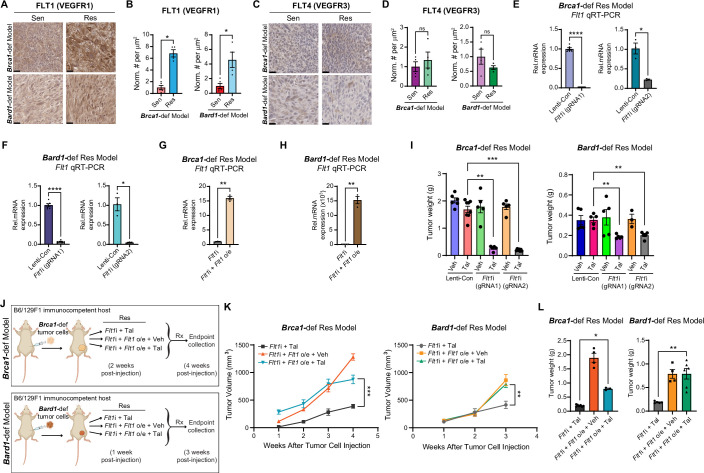
Figure EV3. FLT1 promotes PARPi-resistance in the Brca1-def and Bard1-def breast cancer models.
(A) Representative images of IHC for total FLT1 expression in tumor sections from the mice described in Fig. 1. Scale bars, 20 µm. (B) Immunostained sections from (A) were quantified using automated QuPath software to identify positively stained cells. n = 5 talazoparib-sensitive (“Sen”) tumors, and n = 4 talazoparib-resistant (“Res”) tumors from both Brca1-def and Bard1-def models. Data were presented as mean values ± SEM. P values were determined by a two-tailed, unpaired, Mann–Whitney test. * indicates P = 0.0159 for both models. (C), Representative images of IHC for FLT4 in tumor sections from Fig. 1. Scale bars, 20 µm. (D) Immunostained sections from (C) were quantified using automated QuPath software to identify positively stained cells. n = 4 Sen and Res tumors for both Brca1-def and Bard1-def models. Data were presented as mean values ± SEM. P values were determined by a two-tailed, unpaired, Welch’s test. ns; not significant. (E) qRT-PCR results of Flt1 repression of the indicated groups in the Brca1-def model for both gRNAs. n = 6 (consisting of two independent experiments for each triplicate testing) for both Lenti-Con and Flt1i for gRNA1 and n = 3 (one triplicate testing) for both Lenti-Con and Flt1i for gRNA2. Data were presented as mean values ± SEM. P values were determined by a two-tailed, unpaired, Welch’s test: **** indicates P < 0.0001 for gRNA1 and * indicates 0.0285 for gRNA2. (F) qRT-PCR results of Flt1 repression of the indicated groups in the Bard1-def model. n = 6 (two independent triplicate testing) for both Lenti-Con and Flt1i for gRNA1 and n = 3 (one triplicate testing) for both Lenti-Con and Flt1i for gRNA2. Data were presented as mean values ± SEM. P values were determined by a two-tailed, unpaired, Welch’s test: **** indicates P < 0.0001 for gRNA1 and * indicates P = 0.0273 for gRNA2. (G) qRT-PCR results of Flt1 expression of the indicated groups in the Brca1-def model. n = 3 (one triplicate testing) for Flt1i and Flt1i + Flt1 overexpression (“o/e”) groups. Data were presented as mean values ± SEM. P values were determined by a two-tailed, unpaired, Welch’s test: ** indicates P = 0.0016. (H) qRT-PCR results of Flt1 expression of the indicated groups in the Bard1-def model. n = 3 (one triplicate testing) for Flt1i and Flt1i + Flt1 o/e groups. Data were presented as mean values ± SEM. P values were determined by a two-tailed, unpaired, Welch’s test: ** indicates P = 0.0067. (I) Tumor weights from Fig. 3D were plotted at endpoint. For the Brca1-def model, n = 6 tumors for Lenti-Con + Veh, n = 8 tumors for Lenti-Con + Tal, n = 5 tumors for Flt1i (gRNA1) + Veh or Tal and Flt1i (gRNA2) + Veh, and n = 7 tumors for Flt1i (gRNA2) + Tal treatment groups. For the Bard1-def model, n = 5 tumors for Lenti-Con + Veh or Tal, Flt1i (gRNA1) + Veh or Tal, and Flt1i (gRNA2) + Tal, and n = 3 tumors for Flt1i (gRNA2) + Veh. Data were presented as mean values ± SEM. P values were determined by a two-tailed, unpaired, Mann–Whitney test, comparing endpoint tumor weights between Lenti-Con + Tal and Flt1i (gRNA1 or gRNA2) + Tal groups. For the Brca1-def model, ** indicates P = 0.0016 between Lenti-Con + Tal and Flt1i (gRNA1) + Tal and *** indicates P = 0.0003 between Lenti-Con + Tal and Flt1i (gRNA2) + Tal. For the Bard1-def model, ** indicates P = 0.0079 between Lenti-Con + Tal and Flt1i (gRNA1 or gRNA2) + Tal. (J) Schematic representation of the experiment designed to test whether Flt1 re-expression rescues talazoparib-resistance in Brca1-def and Bard1-def mammary tumors with Flt1 repression. The generation of Brca1- and Bard1-def cancer cells were described in Fig. 3. To stably re-express Flt1, we transduced Flt1-repressed cells with lentiviral particles carrying Flt1 cDNA. For the Brca1-def model, randomized mice received either vehicle (“Veh”) or talazoparib (“Tal”) treatment starting at 2 weeks after tumor-cell injection and were euthanized at 4 weeks following injection. For the Bard1-def model, randomized mice received treatment at 1 week after tumor-cell injection and were euthanized at 3 weeks following injection. (K) Tumor growth curves comparing Flt1i + Tal and Flt1i-Flt1 o/e + Veh or Tal. For the Brca1-def model, n = 7 mice for Flt1i + Tal, n = 4 mice for Flt1i-Flt1 + o/e + Veh, and n = 3 mice for Flt1i-Flt1 + o/e + Tal. For the Bard1-def model, n = 5 mice for Flt1i + Tal, n = 4 mice for Flt1i-Flt1 + o/e + Veh, and n = 6 mice for Flt1i-Flt1 + o/e + Tal. Data were presented as mean values ± SEM. P values were determined with a one-way ANOVA test, comparing endpoint tumor volumes between the Flt1i + Tal and Flt1i-Flt1 + o/e + Tal groups. For the Brca1-def model, *** at 4 weeks indicates P = 0.0001 and for the Bard1-def model, ** at 3 weeks indicates P = 0.0074. (L) Tumor weights from K were plotted at endpoint. For the Brca1-def model, n = 7 tumors for Flt1i + Tal, n = 4 tumors for Flt1i-Flt1 + o/e + Veh, and n = 3 tumors for Flt1i-Flt1 + o/e + Tal. For the Bard1-def model, n = 5 tumors for Flt1i + Tal, n = 4 tumors for Flt1i-Flt1 + o/e + Veh, and n = 6 tumors for Flt1i-Flt1 + o/e + Tal. Data were presented as mean values ± SEM. P values were determined by a two-tailed, unpaired, Mann–Whitney test, comparing endpoint tumor weights between the Flt1i + Tal and Flt1i-Flt1 + o/e + Tal groups. For the Brca1-def model, * indicates P = 0.0167 and for the Bard1-def model, ** indicates P = 0.0043.