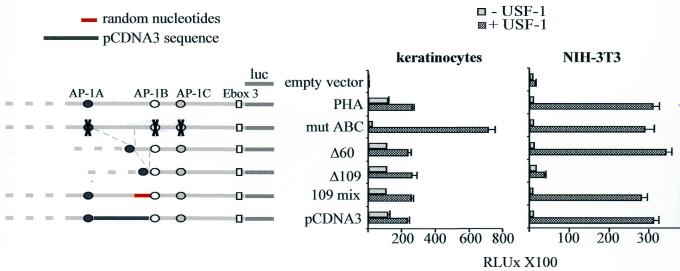
Figure 7.
Functional analysis of the AP-1 binding sites and the intervening sequences within PHA. Full-length, truncated and AP-1-mutated PHA constructs are represented on the left. Keratinocytes and NIH-3T3 cells were transfected with pRK7 USF-1 along with different truncated/mutated constructs derived from PHA. Mut ABC construct corresponds to pHA in which the AP-1 binding sites have been mutated; Δ60 and Δ109 constructs correspond, respectively, to a 60 and 109 bp deletion between the AP1-A and AP1-B binding sites. 109 mix construct represents Δ60 in which 49 bp were inserted in a random order to respect the 109 bp spacing between AP-1 sites. The pCDNA3 construct corresponds to the substitution of 109 nt residing between AP1-A and AP1-B with a 109 bp heterologous sequence from the pCDNA3 polylinker plasmid.