Fig. 2.
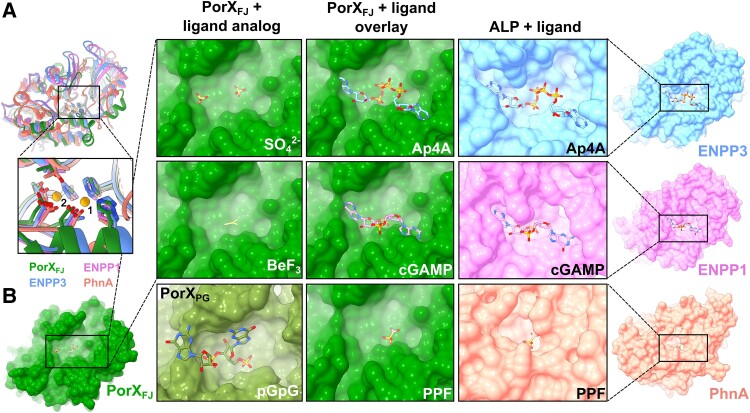
Structural similarities of PorXFJ's APS domain points toward its substrate specificity. A) Overlay of the APS domains of PorXFJ, ENPP3, ENPP1, and PhnA demonstrates their conserved overall fold. Zoom-in view of their catalytic sites including the conserved bi-metal coordination, substrate binding, and catalysis residues. B) Surface representation of APS domains catalytic pockets exhibiting a clear correlation between the ligand and binding pocket dimensions. The APS domain of PorXFJ with its determined ligand analogs (SO42− and BeF3) in green. PorXPG with 5′-phosphoguanylyl-(3′→5′)-guanosine (pGpG) bound in olive green (PDB ID: 7PVK). The ENPP3 with bis(adenosine)-5′-tetraphosphate (Ap4A) polynucleotide bound (PDB ID: 6F2Y) in blue, the ENPP1 with adenosine–guanosine-3′,3′-cyclic monophosphate (cGAMP) bound (PDB ID: 6AEL) in pink, while PhNA with phosphonoformate bound (PDB ID: 1EI6) is in orange. Overlay of PorXFJ with the ligand-bound ENPPs or PhnA suggests that the APS domain of PorXFJ can accommodate a large polyphosphate substrate.