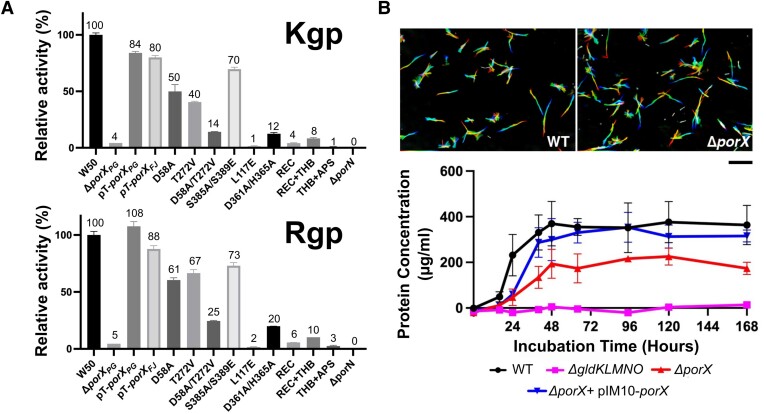
Fig. 6.
In vivo characterization of PorX in P. gingivalis and F. johnsoniae. A) Gingipains enzymatic activity assay of P. gingivalis strain W50 containing different mutants of PorX. W50 and ΔporXPG refer to wild type and PorX null mutant cells, respectively, both carrying empty pT-COW plasmids. The pT-porXPG and pT-porXFJ represent complementation of ΔporXPG pT-COW cells with wild-type PorXPG and PorXFJ, respectively. All other point mutations and truncations include the complementation of ΔporXPG pT-COW cells with PorXPG carrying the relevant mutation. Experiments were performed in quadruplicates, and error bars indicate SDs. B) Top—Gliding motility assay for the wild-type and ΔporX null mutant in F. johnsoniae. A series of 180 frames were collected and colored from red (time zero) to yellow, green, cyan, and finally blue and then integrated into one image, resulting in rainbow traces of gliding cells. Scale bar at lower right indicates 20 μµm and applies to all panels. Bottom—Growth curve of the wild type and ΔporX null mutant F. johnsoniae on a media containing chitin as the sole carbon source. Growth was presented as μg cellular protein/ml. Growth experiments were performed in triplicate, and error bars indicate SDs. The T9SS null mutant, ΔgldKLMNO, that is unable to grow on chitin was used as the negative control. pIM10 contains the wild-type porXFJ and was used to complement the ΔporX mutant.