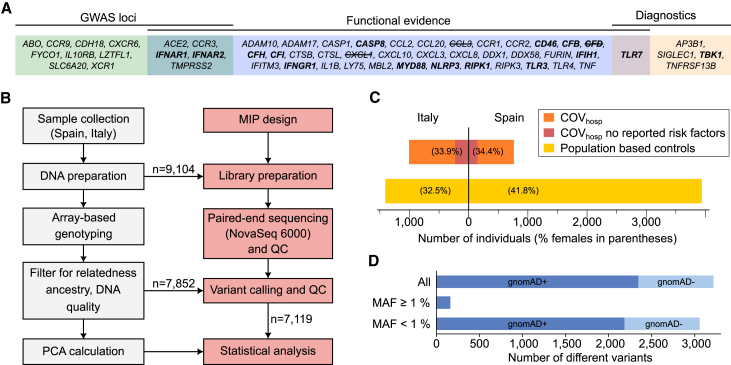
Figure 1.
Study design and cohort characteristics
(A) Candidate genes included in targeted sequencing, grouped according to source of evidence (details in Table S1). Genes known to cause human inborn errors of immunity27 are highlighted in bold, and genes excluded during quality control due to low sequencing coverage are crossed out.
(B) Workflow describing the main steps of sample preparation, genotyping, sequencing, and computational processing. Boxes colored in gray indicate steps that were performed in previous studies.25,26 MIP, molecular inversion probe; PCA, principal-component analysis; QC, quality control.
(C) Number of individuals in the Italian (left) and Spanish (right) subcohorts. The number of COVhosp individuals with no reported risk factors (as described in Table 1) is highlighted in red. The proportion of females is shown in parentheses.
(D) Number of variants observed in the cohort in relation to their minor allele frequency (MAF). In the present study, variants with MAF <1% were denoted as rare variants, while all others were considered common. Intensity of color shading indicates whether (dark) or not (light) variants have been reported with allele frequency in gnomAD r2.1 exomes.