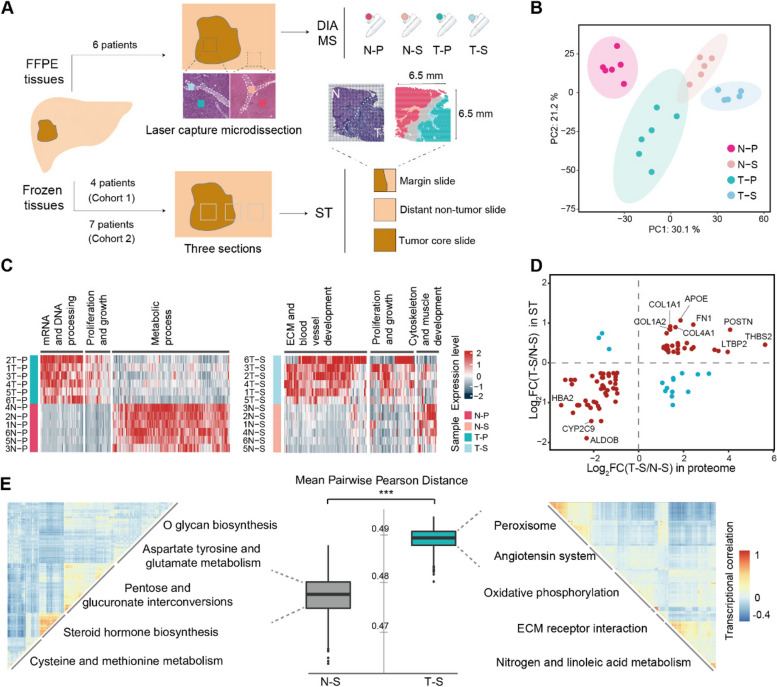
Fig. 1.
Functional diversity and heterogeneity of the tumor stroma in human liver cancer revealed by proteomic and ST analysis. A Tissue processing workflow for proteome and spatial transcriptome. ST, 10X Genomics Visium spatial transcriptomics; DIA-MS, data independent collection-mass spectrometry. B Principal component analysis of protein quantification of all samples. Background ellipses indicate 95% confidence intervals. C Differentially expressed proteins in parenchymal cells (left) and stromal cells (right) in different tissues and their corresponding functions. T–P, tumor parenchymal samples; N–P, non-tumor parenchymal samples; T–S, tumor stroma samples; N‒S, non-tumor stroma samples. D Comparison of the degree of differential gene regulation between T–S and N–S in proteomic and transcriptomic data. Red dots are genes with similar changes between the two datasets, and blue dots are genes with opposite changes. E The heterogeneity of the stromal spots in different areas. The boxplot indicates Pearson’s distance, and heatmaps indicate the similarity of the transcriptional profile, clustered by transcriptional correlation. ***, p < 0.001 by Wilcoxon rank sum test. See also Fig. S1