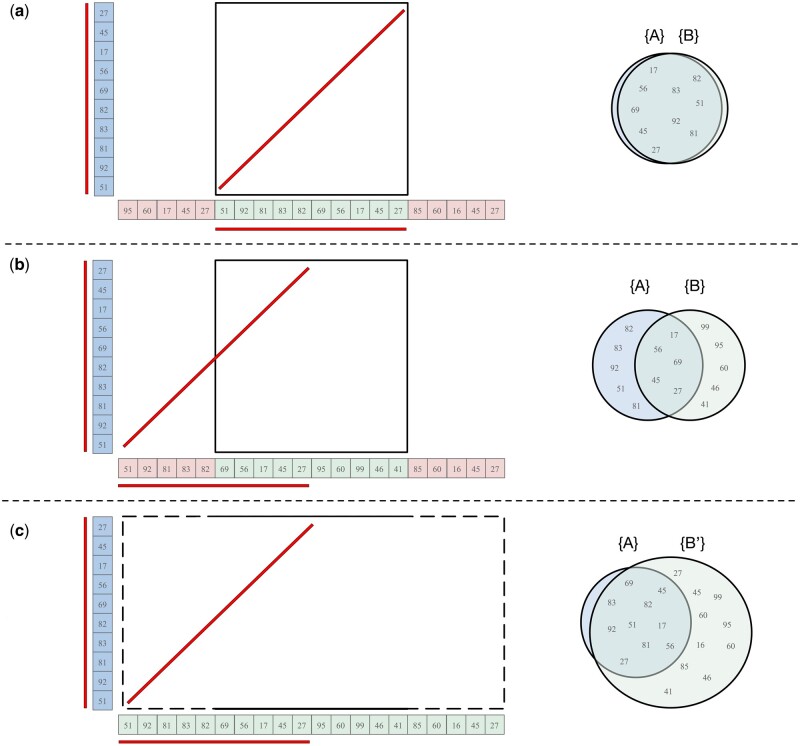
Figure 2.
Sample cases for different interval offsets. k-mers shared between intervals A and B are underlined. (a) In an ideal partition, the shared k-mers are perfectly captured in both intervals. (b) In a worse-case partition, only half of the shared k-mers are captured in the cell, leading to a misleading identity estimate for this region. (c) By keeping A fixed, but expanding B to , ModDotPlot is able to better capture the similarity between two similar sequences with different offsets. The containment index of A in is then used to determine the score of the dot plot matrix cell