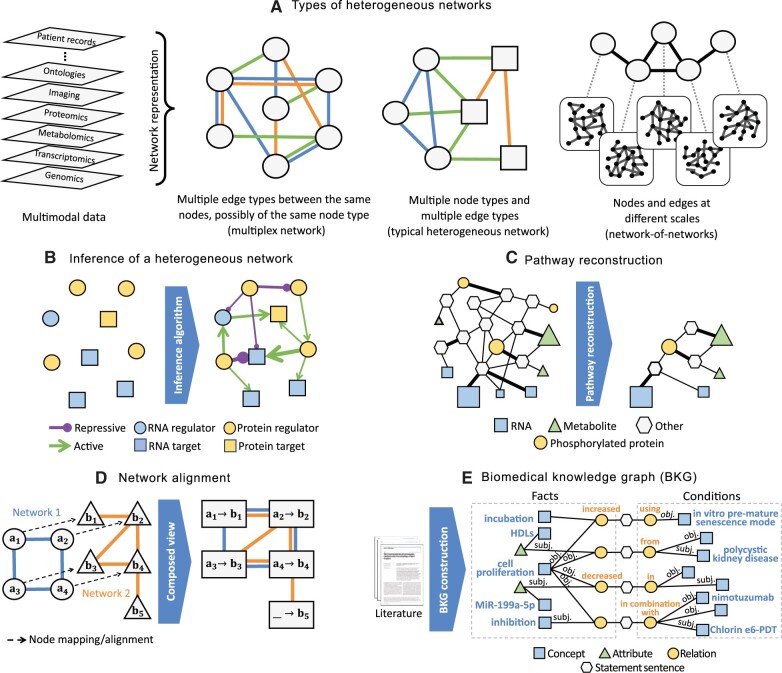
Figure 3.
Prominent topics related to multimodal data integration and heterogeneous networks. (A) Heterogeneous networks can naturally represent multimodal data. A heterogeneous network can have only a single node type, with different data modalities representing multiple edge types. Or, there can exist both multiple node and edge types. Different node types can exist at different biological scales; e.g. in a network-of-networks, nodes at a given scale are networks at the lower scale. (B–E) Prominent topics related to heterogeneous networks. (B) Inference of a heterogeneous network aims to learn the graph topology from multimodal—to date, typically multi-omic—measurements. (C) Pathway reconstruction for interpretation of multi-omic data: the input is multi-omic data and a background molecular network, and the output is a sparse subnetwork. Typically input biomolecules with higher scores (indicated by node sizes) and higher-quality connections (indicated by edge thickness) are prioritized in the output. (D) Network alignment: input can be individual homogeneous networks (left) or heterogeneous networks. Even alignment of homogeneous networks leads to a heterogeneous network (right) whose “supernodes” contain mapped nodes and whose edge types indicate which edges of the original networks are conserved (e.g., between supernodes “a1→b1” and “a2→b2” where the edge exists in both network 1 and network 2) versus nonconserved (e.g. between supernodes “a1→b1” and “a3→b3” where the edge exists in network 1 but not in network 2) under the given node mapping. (E) Inference of and reasoning on BKGs. Shown is a condition-aware BKG. The middle nodes (hexagons) are statement sentences. The layers on their left represent fact tuples and those on their right represent the conditions associated with the facts. The tuples have relation nodes (circles), concept nodes (squares), and optional attribute nodes (triangles).