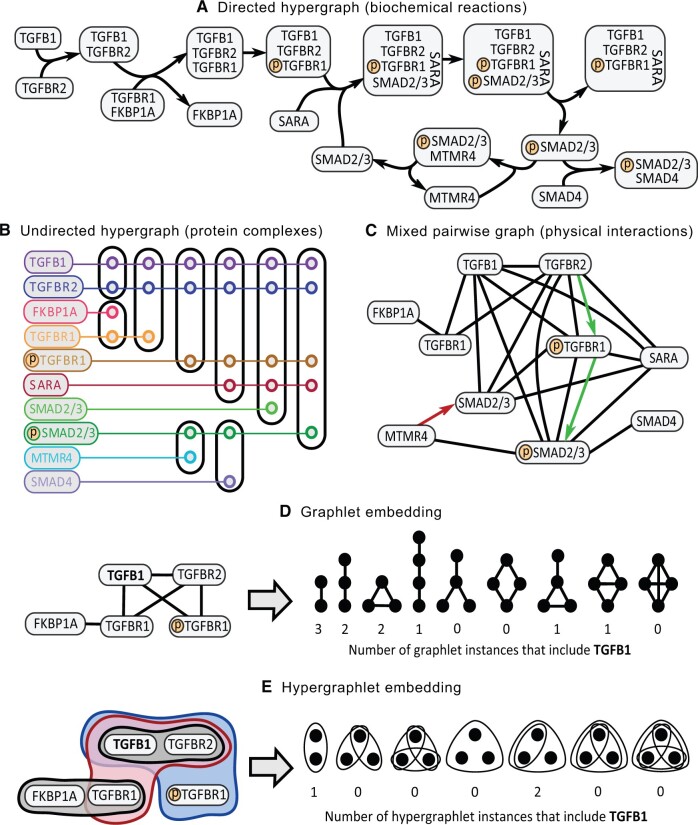
Figure 4.
Graph representations of nine reactions from Reactome’s TGFβ signaling pathway. (A) In a directed hypergraph, each hyperedge captures a reaction (“p” denotes phosphorylation). (B) In an undirected hypergraph, each hyperedge captures a protein complex. (C) In a (mixed) pairwise graph, each edge captures a pairwise interaction. “Mixed” refers to having both directed and undirected edges in the graph. Undirected edges denote physical interactions; directed edges denote either phosphorylation (the two right-most directed edges) or dephosphorylation (the left-most directed edge). (D) A node in a pairwise graph can be represented as a vector of graphlet counts. The number of 2-, 3-, and 4-node graphlet instances that include TGFB1 in the graph on the left are shown. (E) A node in an undirected hypergraph can be represented as a vector of hypergraphlet counts. The number of 2- and 3-node hypergraphlet instances that include TGFB1 in the hypergraph on the left are shown. In panels (D and E), only the (hyper)graphlet-level counts are shown for simplicity, i.e. (hyper)graphlet orbits are not shown nor considered when doing the counting. However, in practice, the more detailed orbit-level counts are computed rather than the (hyper)graphlet-level counts.