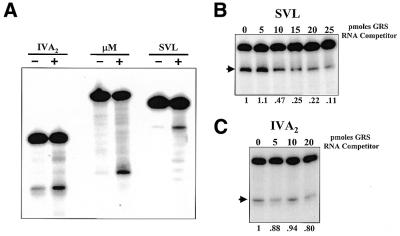
Figure 1.
HnRNP H protein shows signal-specific effects on 3′-end processing. (A) The IVA2, µM and SVL polyadenylation substrate RNAs were incubated in the in vitro 3′-end cleavage system in the absence (– lanes) or presence (+ lanes) of 500 ng recombinant hnRNP H protein. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. The presence of hnRNP H increased the processing efficiency of the IVA2, µM and SVL polyadenylation signals by 3.7 ± 0.8-, 3.9 ± 0.8- and 3.6 ± 0.5-fold, respectively. (B) The SVL polyadenylation substrate RNA was incubated in the in vitro 3′-end cleavage system in the presence of the indicated amounts of a synthetic competitor RNA, GRS, that can bind and sequester hnRNP H protein. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. The processing efficiency relative to 0 pmol GRS RNA competitor is shown below each lane. (C) The IVA2 polyadenylation substrate RNA was incubated in the in vitro 3′-end cleavage system in the presence of the indicated amounts of synthetic GRS competitor RNA. Reaction products were analyzed on a 5% acrylamide gel containing 7 M urea. The processing efficiency relative to 0 pmol GRS RNA competitor is shown below each lane. The arrows on the left of (B) and (C) indicate the positions of the 5′ cleavage product.