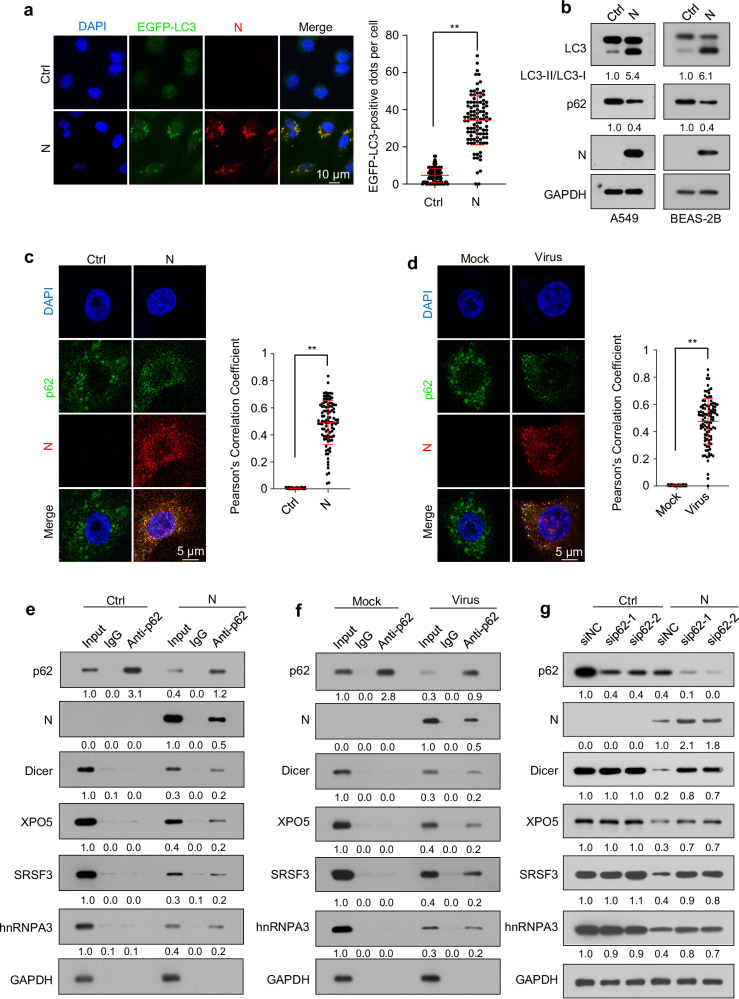
Fig. 2. SARS-CoV-2 N protein mediates the interactions of Dicer, XPO5, SRSF3, and hnRNPA3 with p62 and induces their autophagic degradation.
a The formation of EGFP-LC3 puncta in A549 cells stably expressing EGFP-LC3 transfected with N protein-expressing plasmid or control plasmid (n = 100 cells from three biological replicates). b LC3-II and p62 levels in A549-Ctrl and A549-N cells (left) or BEAS-2B-Ctrl and BEAS-2B-N cells (right). c Immunofluorescence of A549-Ctrl or A549-N cells treated with chloroquine using anti-p62 and anti-N protein antibodies and DAPI for nuclear staining. d Immunofluorescence of A549-hACE2 cells infected with SARS-CoV-2 or mock-infected and treated with chloroquine using anti-p62 and anti-N protein antibodies and DAPI for nuclear staining. Pearson′s correlation coefficient of the colocalization between N protein and p62 in (c, d) (n = 100 cells from three biological replicates). e Cell lysates of A549-Ctrl and A549-N cells were subjected to immunoprecipitation using an anti-p62 antibody and immunoblotting with the indicated antibodies. f Cell lysates of A549-hACE2 cells infected with SARS-CoV-2 or mock were subjected to immunoprecipitation using an anti-p62 antibody and immunoblotting with the indicated antibodies. g Immunoblotting analysis of the indicated proteins in A549-Ctrl and A549-N cells transfected with a negative control siRNA (siNC) or p62-specific (sip62) siRNAs. The numbers below the blots indicate the relative densitometric quantification of the bands normalized to the corresponding GAPDH band (b, g) or input (e, f); the mean values of three independent experiments are shown. Data in (a, c, d) are expressed as mean ± SD. **p < 0.01 (two-tailed unpaired Student’s t-test). Ctrl control plasmid, N N protein, anti-N anti-N protein antibody. Source data and exact p values are provided in the Source Data file.