Abstract
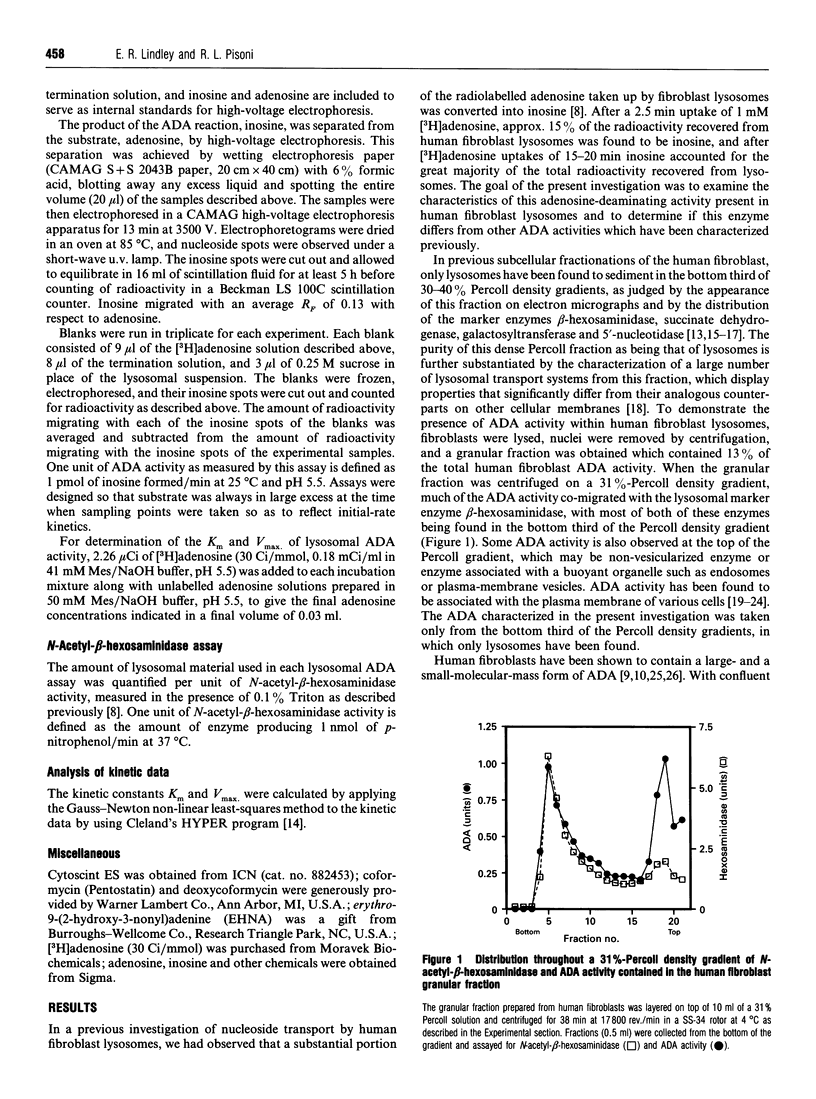
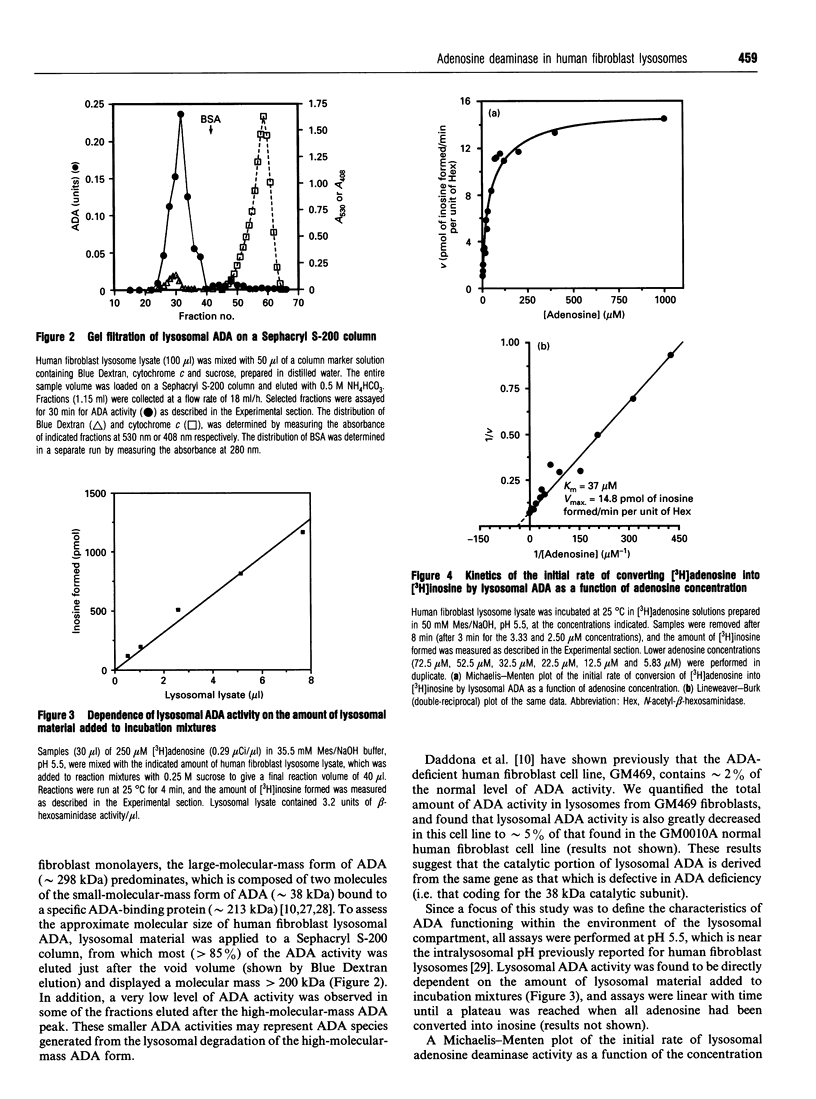
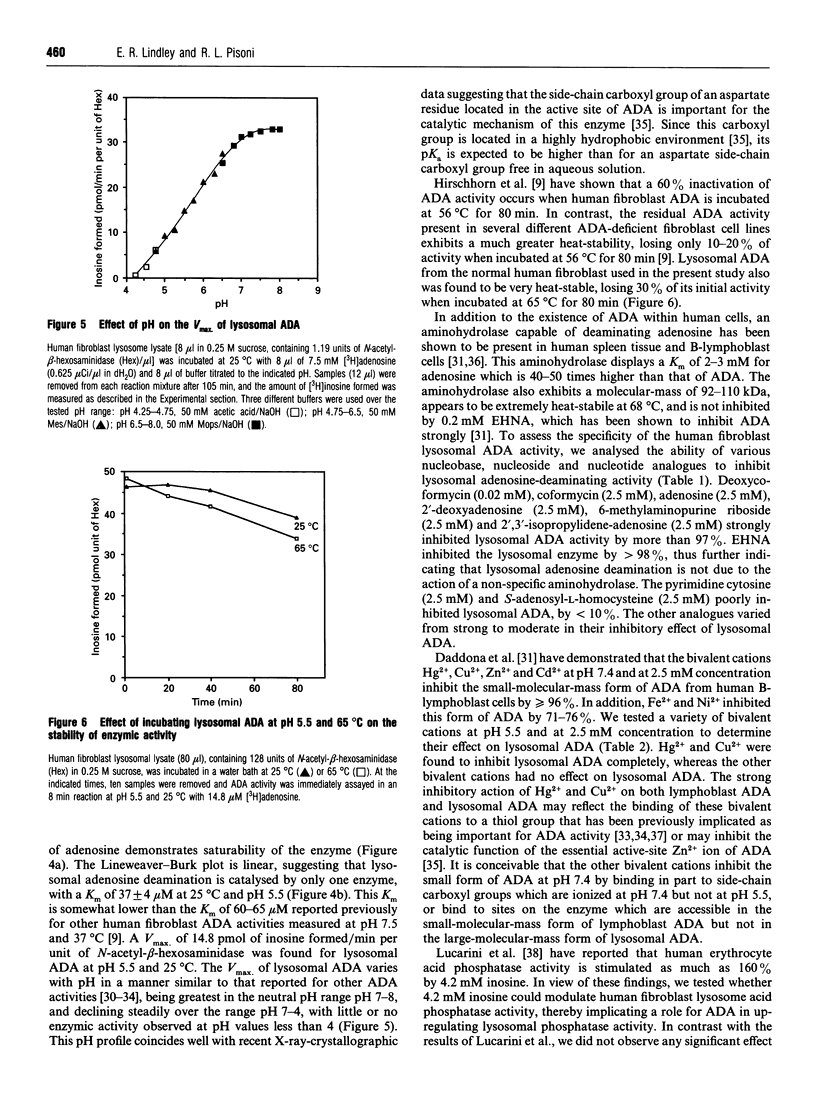
Human fibroblast lysosomes, purified on Percoll density gradients, contain an adenosine deaminase (ADA) activity that accounts for approximately 10% of the total ADA activity in GM0010A human fibroblasts. In assays of lysosomal ADA, the conversion of [3H]adenosine into [3H]inosine was proportional to incubation time and the amount of lysosomal material added to reaction mixtures. Maximal activity was observed between pH 7 and 8, and lysosomal ADA displayed a Km of 37 microM for adenosine at 25 degrees C and pH 5.5. Lysosomal ADA was completely inhibited by 2.5 mM Cu2+ or Hg2+ salts, but not by other bivalent cations (Ba2+, Cd2+, Ca2+, Fe2+, Mg2+, Mn2+ and Zn2+). Coformycin (2.5 mM), deoxycoformycin (0.02 mM), 2'-deoxyadenosine (2.5 mM), 6-methylaminopurine riboside (2.5 mM), 2'-3'-isopropylidene-adenosine (2.5 mM) and erythro-9-(2-hydroxy-3-nonyl)adenine (0.2 mM) inhibited lysosomal ADA by > 97%. In contrast, 2.5 mM S-adenosyl-L-homocysteine and cytosine were poor inhibitors. Nearly all lysosomal ADA activity is eluted as a high-molecular-mass protein (> 200 kDa) just after the void volume on a Sephacryl S-200 column, and is very heat-stable, retaining 70% of its activity after incubation at 65 degrees C for 80 min. We speculate that compartmentalization of ADA within lysosomes would allow deamination of adenosine to occur without competition by adenosine kinase, which could assist in maintaining cellular energy requirements under conditions of nutritional deprivation.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akedo H., Nishihara H., Shinkai K., Komatsu K., Ishikawa S. Multiple forms of human adenosine deaminase. I. Purification and characterization of two molecular species. Biochim Biophys Acta. 1972 Jul 13;276(1):257–271. doi: 10.1016/0005-2744(72)90028-9. [DOI] [PubMed] [Google Scholar]
- Andy R. J., Kornfeld R. The adenosine deaminase binding protein of human skin fibroblasts is located on the cell surface. J Biol Chem. 1982 Jul 25;257(14):7922–7925. [PubMed] [Google Scholar]
- Aran J. M., Colomer D., Matutes E., Vives-Corrons J. L., Franco R. Presence of adenosine deaminase on the surface of mononuclear blood cells: immunochemical localization using light and electron microscopy. J Histochem Cytochem. 1991 Aug;39(8):1001–1008. doi: 10.1177/39.8.1856451. [DOI] [PubMed] [Google Scholar]
- Arsenis C., Gordon J. S., Touster O. Degradation of nucleic acids by lysosomal extracts of rat liver and Ehrlich ascites tumor cells. J Biol Chem. 1970 Jan 10;245(1):205–211. [PubMed] [Google Scholar]
- Arsenis C., Touster O. Purification and properties of an acid nucleotidase from rat liver lysosomes. J Biol Chem. 1968 Nov 10;243(21):5702–5708. [PubMed] [Google Scholar]
- Bernardi G. Mechanism of action and structure of acid deoxyribonuclease. Adv Enzymol Relat Areas Mol Biol. 1968;31:1–49. doi: 10.1002/9780470122761.ch1. [DOI] [PubMed] [Google Scholar]
- Bielat K., Tritsch G. L. Ecto-enzyme activity of human erythrocyte adenosine deaminase. Mol Cell Biochem. 1989 Apr 11;86(2):135–142. doi: 10.1007/BF00222613. [DOI] [PubMed] [Google Scholar]
- Cleland W. W. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- Daddona P. E., Frohman M. A., Kelley W. N. Human adenosine deaminase and its binding protein in normal and adenosine deaminase-deficient fibroblast cell strains. J Biol Chem. 1980 Jun 25;255(12):5681–5687. [PubMed] [Google Scholar]
- Daddona P. E., Kelley W. N. Characteristics of an aminohydrolase distinct from adenosine deaminase in cultured human lymphoblasts. Biochim Biophys Acta. 1981 Apr 14;658(2):280–290. doi: 10.1016/0005-2744(81)90298-9. [DOI] [PubMed] [Google Scholar]
- Daddona P. E., Kelley W. N. Human adenosine deaminase. Purification and subunit structure. J Biol Chem. 1977 Jan 10;252(1):110–115. [PubMed] [Google Scholar]
- Daddona P. E., Kelley W. N. Human adenosine deaminase. Stoichiometry of the adenosine deaminase-binding protein complex. Biochim Biophys Acta. 1979 Oct 24;580(2):302–311. doi: 10.1016/0005-2795(79)90143-0. [DOI] [PubMed] [Google Scholar]
- Fox I. H., Kelley W. N. The role of adenosine and 2'-deoxyadenosine in mammalian cells. Annu Rev Biochem. 1978;47:655–686. doi: 10.1146/annurev.bi.47.070178.003255. [DOI] [PubMed] [Google Scholar]
- Franco R., Aran J. M., Colomer D., Matutes E., Vives-Corrons J. L. Association of adenosine deaminase with erythrocyte and platelet plasma membrane: an immunological study using light and electron microscopy. J Histochem Cytochem. 1990 May;38(5):653–658. doi: 10.1177/38.5.2332624. [DOI] [PubMed] [Google Scholar]
- Franco R., Centelles J. J. Release of adenosine deaminase from rat intestinal mucosa. Biochem Soc Trans. 1990 Aug;18(4):641–642. doi: 10.1042/bst0180641. [DOI] [PubMed] [Google Scholar]
- Gieselmann V., Pohlmann R., Hasilik A., Von Figura K. Biosynthesis and transport of cathepsin D in cultured human fibroblasts. J Cell Biol. 1983 Jul;97(1):1–5. doi: 10.1083/jcb.97.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydrick S. J., Lardeux B. R., Mortimore G. E. Uptake and degradation of cytoplasmic RNA by hepatic lysosomes. Quantitative relationship to RNA turnover. J Biol Chem. 1991 May 15;266(14):8790–8796. [PubMed] [Google Scholar]
- Hirschhorn R., Beratis N. G., Martiniuk F. Adenosine deaminase. Alterations in activity and isozymes during growth of normal and genetically deficient fibroblasts. Exp Cell Res. 1978 Nov;117(1):103–109. doi: 10.1016/0014-4827(78)90432-9. [DOI] [PubMed] [Google Scholar]
- Hirschhorn R., Beratis N., Rosen F. S. Characterization of residual enzyme activity in fibroblasts from patients with adenosine deaminase deficiency and combined immunodeficiency: evidence for a mutant enzyme. Proc Natl Acad Sci U S A. 1976 Jan;73(1):213–217. doi: 10.1073/pnas.73.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inwood M., Povey S., Delhanty J. D. Comparison of isozymes in fetal, adult and transformed fibroblasts. Cell Biol Int Rep. 1980 Apr;4(4):327–335. doi: 10.1016/0309-1651(80)90214-3. [DOI] [PubMed] [Google Scholar]
- Jarvis S. M., Young J. D., Ansay M., Archibald A. L., Harkness R. A., Simmonds R. J. Is inosine the physiological energy source of pig erythrocytes? Biochim Biophys Acta. 1980 Mar 27;597(1):183–188. doi: 10.1016/0005-2736(80)90162-5. [DOI] [PubMed] [Google Scholar]
- Kipshidze N. N., Karsanov N. V., Chichinadze N. A., Machitadze T. N., Azrumelashvili M. I. Inosine effect on high-energy phosphate content, mechanical activity of glycerinated fiber bundles, morphology and microcirculation in myocardium in toxi-allergic and allergic myocarditis. Methods Find Exp Clin Pharmacol. 1983;5(3):163–170. [PubMed] [Google Scholar]
- Kovacević Z., Popović J., Brkljac O., Lelas S. Interaction of metabolism of aspartate and inosine and energy state of malignant cells. Biochem J. 1987 Oct 1;247(1):47–51. doi: 10.1042/bj2470047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardeux B. R., Mortimore G. E. Amino acid and hormonal control of macromolecular turnover in perfused rat liver. Evidence for selective autophagy. J Biol Chem. 1987 Oct 25;262(30):14514–14519. [PubMed] [Google Scholar]
- Lucarini N., Borgiani P., Ballarini P., Bottini E. Erythrocyte acid phosphatase (ACP1) activity. In vitro modulation by adenosine and inosine and effects of adenosine deaminase (ADA) polymorphism. Hum Genet. 1989 Jan;81(2):185–187. doi: 10.1007/BF00293900. [DOI] [PubMed] [Google Scholar]
- Orsi B. A., McFerran N., Hill A., Bingham A. Kinetics and the mechanism of action of adenosine aminohydrolase. Biochemistry. 1972 Aug 29;11(18):3386–3392. doi: 10.1021/bi00768a011. [DOI] [PubMed] [Google Scholar]
- Pertoft H., Wärmegård B., Hök M. Heterogeneity of lysosomes originating from rat liver parenchymal cells. Metabolic relationship of subpopulations separated by density-gradient centrifugation. Biochem J. 1978 Jul 15;174(1):309–317. doi: 10.1042/bj1740309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrogner N. Adenosine deaminase from calf spleen. II. Chemical and enzymological properties. Arch Biochem Biophys. 1967 Mar;119(1):147–154. doi: 10.1016/0003-9861(67)90440-7. [DOI] [PubMed] [Google Scholar]
- Pisoni R. L. Characterization of a phosphate transport system in human fibroblast lysosomes. J Biol Chem. 1991 Jan 15;266(2):979–985. [PubMed] [Google Scholar]
- Pisoni R. L., Flickinger K. S., Thoene J. G., Christensen H. N. Characterization of carrier-mediated transport systems for small neutral amino acids in human fibroblast lysosomes. J Biol Chem. 1987 May 5;262(13):6010–6017. [PubMed] [Google Scholar]
- Pisoni R. L., Thoene J. G. Detection and characterization of a nucleoside transport system in human fibroblast lysosomes. J Biol Chem. 1989 Mar 25;264(9):4850–4856. [PubMed] [Google Scholar]
- Pisoni R. L., Thoene J. G., Lemons R. M., Christensen H. N. Important differences in cationic amino acid transport by lysosomal system c and system y+ of the human fibroblast. J Biol Chem. 1987 Nov 5;262(31):15011–15018. [PubMed] [Google Scholar]
- Pisoni R. L., Thoene J. G. The transport systems of mammalian lysosomes. Biochim Biophys Acta. 1991 Dec 12;1071(4):351–373. doi: 10.1016/0304-4157(91)90002-e. [DOI] [PubMed] [Google Scholar]
- Porat N., Gill D., Parola A. H. Adenosine deaminase in cell transformation. Biophysical manifestation of membrane dynamics. J Biol Chem. 1988 Oct 15;263(29):14608–14611. [PubMed] [Google Scholar]
- Rome L. H., Garvin A. J., Allietta M. M., Neufeld E. F. Two species of lysosomal organelles in cultured human fibroblasts. Cell. 1979 May;17(1):143–153. doi: 10.1016/0092-8674(79)90302-7. [DOI] [PubMed] [Google Scholar]
- Ronca G., Bauer C., Rossi C. A. Role of sulphydryl groups in adenosine deaminase. Eur J Biochem. 1967 Jun;1(4):434–438. doi: 10.1007/978-3-662-25813-2_59. [DOI] [PubMed] [Google Scholar]
- Schrader W. P., Pollara B., Meuwissen H. J. Characterization of the residual adenosine deaminating activity in the spleen of a patient with combined immunodeficiency disease and adenosine deaminase deficiency. Proc Natl Acad Sci U S A. 1978 Jan;75(1):446–450. doi: 10.1073/pnas.75.1.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. L., Greene A. A., Potashnik R., Mendoza S. A., Schneider J. A. Lysosomal cystine transport. Effect of intralysosomal pH and membrane potential. J Biol Chem. 1987 Jan 25;262(3):1244–1253. [PubMed] [Google Scholar]
- Swallow D. M., Evans L., Hopkinson D. A. Several of the adenosine deaminase isozymes are glycoproteins. Nature. 1977 Sep 15;269(5625):261–262. doi: 10.1038/269261a0. [DOI] [PubMed] [Google Scholar]
- Van der Weyden M. B., Kelley W. N. Human adenosine deaminase. Distribution and properties. J Biol Chem. 1976 Sep 25;251(18):5448–5456. [PubMed] [Google Scholar]
- Wilson D. K., Rudolph F. B., Quiocho F. A. Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science. 1991 May 31;252(5010):1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- Yoshiyama M., Sakai H., Teragaki M., Takeuchi K., Takeda T., Ikata M., Ishikawa M., Miura I. The effect of inosine on the post ischemic heart as bio-energy recovering factor in 31P-MRS. Biochem Biophys Res Commun. 1988 Mar 30;151(3):1408–1415. doi: 10.1016/s0006-291x(88)80519-9. [DOI] [PubMed] [Google Scholar]
- Young J. D., Jarvis S. M., Clanachan A. S., Henderson J. F., Paterson A. R. Nitrobenzylthioinosine: an in vivo inhibitor of pig erythrocyte energy metabolism. Am J Physiol. 1986 Jul;251(1 Pt 1):C90–C94. doi: 10.1152/ajpcell.1986.251.1.C90. [DOI] [PubMed] [Google Scholar]
- Young J. D., Paterson A. R., Henderson J. F. Nucleoside transport and metabolism in erythrocytes from the Yucatan miniature pig. Evidence that inosine functions as an in vivo energy substrate. Biochim Biophys Acta. 1985 Oct 17;842(2-3):214–224. doi: 10.1016/0304-4165(85)90205-3. [DOI] [PubMed] [Google Scholar]
- Zaporowska-Siwiak E., Michalik M., Kajstura J., Korohoda W. Density-dependent survival of Ehrlich ascites tumour cells in the presence of various substrates for energy metabolism. J Cell Sci. 1985 Aug;77:75–85. doi: 10.1242/jcs.77.1.75. [DOI] [PubMed] [Google Scholar]
- Zeidler R. B., Metzler M. H., Moran J. B., Kim H. D. The liver is an organ site for the release of inosine metabolized by non-glycolytic pig red cells. Biochim Biophys Acta. 1985 Mar 8;838(3):321–328. doi: 10.1016/0304-4165(85)90229-6. [DOI] [PubMed] [Google Scholar]


