Abstract
Laboratory assays that can accurately distinguish recent (occurring within the past year) from long-standing (>1 year) HIV infection are crucial for understanding HIV transmission dynamics in a population. However, often these efforts are confounded by inaccurate HIV diagnosis and the presence of HIV-2 in the population being surveyed. This study describes development of a multiplex assay that can simultaneously perform HIV diagnosis, HIV serotyping, and detection of recent HIV-1 infection in a single well. HIV diagnosis and HIV-2 serotyping were accomplished by coupling beads with an HIV-1 p24-gp41 fusion protein and HIV-2 peptide from gp36 immunodominant region, respectively. HIV-1 recent infection detection was accomplished by coupling beads with limiting amounts of multi-subtype gp41 immunodominant protein, recombinant immunodominant region, group M (rIDR-M). Assay conditions, including concentration of coupled antigens, were systematically optimized using well-characterized specimens with known HIV-status (positive or negative), HIV-2 specimens, and recent or long-term HIV-1 classification based on LAg-Avidity enzyme immunoassay (EIA) in a stepwise manner. Beads were then combined in a multiplex assay to evaluate its performance using large panel of specimens (n = 1,500) that included HIV-1 positive (n = 570, recent = 78, long-term = 492), HIV-2 positive (n = 31), and seronegative individuals (n = 899). The diagnostic component of the assay performed with high sensitivity (99.8%) and specificity (99.7%), while the HIV-2 serotyping sensitivity and specificity were 96.7% and 100%, respectively. There was a high correlation (R = 0.84) between the LAg-Avidity EIA and the multiplex assay for recent infection detection. The assay showed high inter- and intra-assay reproducibility with %coefficient of variation of <10% in the dynamic range. The multiplex assay has the ability to diagnose HIV infection, perform serotyping, and detect and distinguish recent from long-term HIV infections, all in a single well. This novel assay has the potential to simplify HIV surveillance by reducing the multiple steps that are otherwise required.
Keywords: HIV, prevalence, incidence, LAg-Avidity EIA, multiplex, serotyping
Introduction
With estimated 37 million HIV-infected individuals worldwide,1 and in the absence of an effective vaccine, the prevention of HIV transmission remains an essential goal for epidemic control. Accurate determination of HIV-1 incidence, which is the rate of new infections in a population, is critical in tracking the leading edge of the epidemic. Current methods used in determining HIV-1 incidence have many limitations. These methods include mathematical modeling, which requires numerous assumptions and inputs and may pose additional challenges in the era of expanding antiretroviral treatment (ART) and longer survival of HIV-1-infected individuals. On the other hand prospective longitudinal cohort studies can be used to estimate incidence; however, they often times are labor-intensive, complex, very expensive, and have recruitment biases.
Recently, CD4 distribution levels in newly diagnosed persons have been used to estimate population incidence using CD4 depletion model in case-based surveillance in the United States.2 Although this assumes that CD4 depletion is similar in different population infected with different subtypes,2 Moreover, this approach requires robust case-based surveillance with weighted probabilities assigned to each person found to be HIV-positive based on number of assumptions. Laboratory methods can be unbiased and do not require complicated assumptions and weighting to each case. Therefore, the development of laboratory-based assays has gained traction because of their simplicity, low cost, and the application to cross-sectional specimens.
Efforts to detect recent HIV-1 infection using laboratory assays started in the mid-1990s. In the late 1990, several advancements were made in serological assays that made use of both antibody titers and avidity after seroconversion to distinguish between recent and long-term infection.3-10 This period began with a seminal article published on the detuned assay: a modification of the conventional ELISA to allow discrimination based on antibody titer.10 More recently, progress has been made in the development of novel assays and the identification of new biomarkers for HIV-1 incidence.11-15 The BED-Capture enzyme immunoassay (BED-CEIA) and the Limiting Antigen Avidity EIA (LAg-Avidity EIA) were developed and commercialized for surveillance use to estimate HIV-1 incidence in the population16-22 and have been widely used in surveys. The LAg-Avidity EIA uses a multi-subtype gp41 protein, recombinant immunodominant region, group M (rIDR-M) covering the immunodominant region of gp41 of HIV-1 and a novel approach to measuring antibody avidity using limiting amount of antigen.15
The LAg-Avidity EIA and other antibody-based incidence assays are performed only on confirmed HIV-1 positive specimens because these assays are designed to identify recently infected persons post-seroconversion. These assays cannot be used on specimens that are HIV-1 false positive (FP) or HIV-2 positive because these assays are designed for confirmed HIV-1 positive cases. All FP or HIV-2 specimens, if any, will test recent and will elevate incidence. Therefore, prior accurate diagnosis of HIV-1 and identification of HIV-2, if any, is very important. Moreover, the presence of false-positives is not uncommon in surveillance settings. Therefore, LAg incidence testing algorithm requires serology confirmation of specimens that are below certain threshold. Although the use of additional diagnostic testing (serology confirmation and HIV-2 diagnosis) has led to accurate incidence estimates when properly executed and weighted, this requires the use of multiple assays, inherently additional sample volume, and several different instruments resulting in increased cost and labor. The compounded errors resulting from many technical steps, if not executed properly, are often not desirable.
Traditionally, ELISA-based assays have been used to measure antibody avidity and antibody titers. In each case, the assay is directed against a single analyte or a single parameter, such that measuring multiple parameters would require multiple plates or wells.3 Multiplexing allows for the analysis of greater number of analytes in a single well. Bead-based multiplex assays are gaining traction in the diagnosis of infectious diseases.23-26 Each bead is color-coded with a unique spectral property. Colored beads are coupled with different capture agents and incubated with samples. A detection antibody, which is conjugated to a fluorophore, is then used to quantitatively visualize the captured analyte, at the same time that the beads are identified by their spectral properties.
A multiplex assay has been previously described recently that utilizes multiple biomarkers, and recent infection is determined by an individual’s antibody reactivity profile.12 However, this approach is complex and fails to address the key contributing factors to elevated incidence in surveys (i.e., FPs and HIV-2). Limitations with current assays are not principally associated with the use of a single analyte, but rather subtype diversity, misdiagnosis of HIV-1 infection, improper serotyping, and the use of ART.13,15,27,28
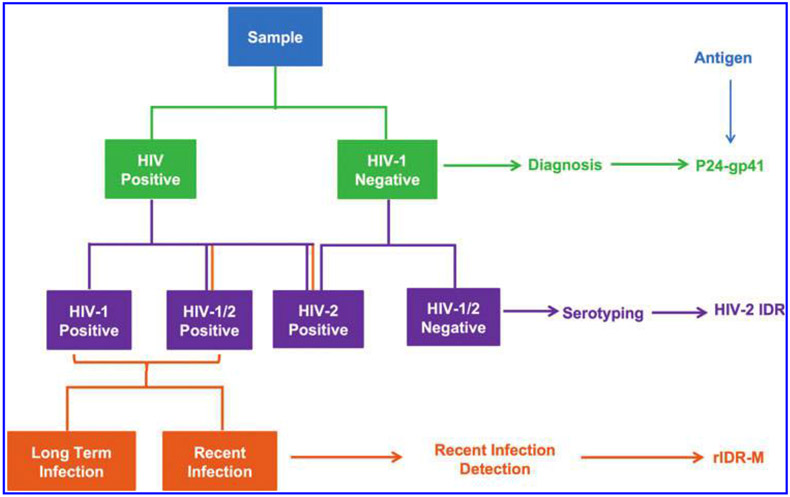
In this study, we combined the principle of HIV diagnostics and serotyping, concept of limiting antigen, and power of multiplexing to develop a multiplex assay with the ability to detect and differentiate antibodies to HIV-1 and HIV-2 and distinguish between recent and long-term infections from samples of diverse origins (Fig. 1). We have described here the development of the assay and its performance characteristics. We have compared these results with the combined results from multiple HIV diagnostic tests, serotyping, and detection of recent infection.
FIG. 1.
Schematic representation of the multiplex assay. The scheme shows the serological parameters and logical approach that was used for diagnostics, serotyping, and incident infection determination and the antigen used for each parameter.
Materials and Methods
Specimens
Plasma specimens for this study were from our specimen bank collected over a period of last 10 years and were derived from various sources. This included rejected units from blood banks collected from multiple countries to develop a worldwide panel and additional commercially available bulk volume specimens with HIV-1 antibodies, HIV-2 antibodies, or seronegatives. The specimens were thoroughly characterized using a battery of tests, aliquoted, and stored frozen at −20°C or below. Stored plasma samples were categorized in panels as illustrated in Table 1. All specimens were characterized by third generation Genetic Systems EIA (Bio-Rad Laboratory, Hercules, CA) followed by western blot assay (Maxim, Rockville, MD). HIV-1 and HIV-2 serotyping was done using Multispot HIV-1/2 (from Bio-Rad). Additional characterization of HIV-1 specimens was done using LAg-Avidity EIA (Sedia BioSciences, Portland, OR) to classify specimens as recent or long-term infections. These data were used for comparison and for assessing performance of the multiplex assay. Panels 1 and 3 were made available from commercial sources, while some specimens from panel 2 are a subset of panel 5. Panel composition changed from simple (panel 1) to increasing complexity (panel 5) to allow stepwise development and characterization of the multiplex assay.
Table 1.
Specimen Panels Used for the Development, Optimization, and Evaluation of the Multiplex Assay
| Specimen | Total no. | HIV-1 or dual | HIV-2 | Negatives | Purpose |
|---|---|---|---|---|---|
| Panel 1 | 10 | 5 | 0 | 5 | HIV-1 diagnostic optimization |
| Panel 2 | 13 | 4 | 7 | 2 | Serotyping optimization |
| Panel 3 | 15 | 15* | 0 | 0 | Incidence optimization |
| Panel 4 | 85 | 37* | 3 | 45 | Small panel combined evaluation |
| Panel 5 | 1,500 | 570* | 31 | 899 | Large panel combined evaluation |
Specimens are obtained from different countries (Kenya, Ivory Coast, Thailand, Uganda, United States, Cameroon, and South Africa) and commercially from Boca Biolistics. These specimens are well characterized using HIV ELISA and Western blot (for diagnosis), Multispot (for HIV-2) and LAg-Avidity EIA (for recency classification).
represents specimens that are further classified as HIV-1 recent or HIV-1 long-term by LAg-Avidity EIA.
Panel 1.
This panel was comprised of well-characterized HIV-1 positive (n = 5) and negative (n = 5) specimens that have been primarily used in our laboratory for HIV-1 competency testing and training. Panel 1 specimens were used in the optimization of the diagnostic component of the multiplex assay.
Panel 2.
This was a 13-member panel containing well-characterized HIV-1 (n = 4), HIV-2 (n = 7), and negatives (n = 2) specimens. Panel 2 specimens were used in the optimization of the serotyping component of the multiplex assay to distinguish HIV-1 and HIV-2.
Panel 3.
This panel contained 15 standard HIV-1 positive specimens (8 recent and 7 long-term) generally used for the training of the LAg-Avidity EIA and also in the development of new incidence assays. Panel 3 specimens were used in the optimization of the incidence component of the multiplex assay.
Panel 4.
Panel 4 consisted of commercially acquired bulk volume specimens from Boca Biolistics, Inc. (Coconut Creek, FL). This 85-member panel contains HIV-1 (n = 35), HIV-2 (n = 3), HIV-1/2 (n = 2) positive specimens and negative specimens (n = 45). HIV-1 specimens included 26 recent and 11 long-term infection as classified by the LAg-Avidity EIA. Panel 4 specimens were used in the small-scale evaluation of the performance of the multiplex assay to simultaneously diagnose, perform serotyping, and identify recent infections. It was also used to determine reproducibility parameters of the assay and compare the mean fluorescent intensity (MFI) from each antigen in the multiplex and monoplex formats.
Panel 5.
This 1,500-member panel contained well-characterized HIV-1 positive (n = 570), HIV-2 positive (n = 31) and negative specimens (n = 899). HIV-1 specimens include 78 recent and 492 long-term infection as classified by the LAg-Avidity EIA. These specimens were obtained from different countries: Cameroon (n = 56), Kenya (n = 475), Ivory Coast (n = 361), Thailand (n = 199), Uganda (n = 27), United States (n = 197), South Africa (n = 165), Sierra Leone (n = 20); and very likely represented different subtypes and recombinants due to varying geographic locations. Specimens from Sierra Leone were commercially purchased from Boca Biolistics, Inc. These 1,500 specimens are routinely used for the validation of new HIV diagnostic tests. In this assay, panel 5 specimens were used in a large-scale evaluation of the performance of the multiplex assay to simultaneously diagnose, perform serotyping, and identify recent infections.
Coupling of HIV antigens to beads
Diagnostic antigen.
The p24-gp41 fusion protein (Bio-process, Inc., Australia) was coupled at saturating concentration (1 μg/1.5 million beads) on bead region 12 (Table 2). The lyophilized p24-gp41 fusion protein was dissolved in coupling buffer at high concentration (0.44 mg/mL) and stored in 20 μL aliquots at −80°C.
Table 2.
Critical Assay Parameter Showing the Antigen Used, Range of Antigen Concentrations Tested, and the Optimal Antigen Concentration Experimentally Determined
| Parameter | Purpose | Range tested | Optimum concentration |
|---|---|---|---|
| p24-gp41 Protein | Diagnostic antigen | 50 μg/1.5 M beads to 0.25 μg/1.5 M beads | 1 μg/1.5 M beads |
| HIV-2 IDR peptide | Serotyping antigen | 20 μg/1.5 M beads to 0.5 μg/1.5 M beads | 10 μg/1.5 M beads |
| rIDR-M | Recent and long term separation antigen | 1 μg/1.5 M beads to 0.025 μg/1.5 M beads | 0.04 μg/1.5 M beads |
Serotyping antigen.
The HIV-2 IDR peptide, which is derived from the immunodominant region of HIV-2 gp36, has been previously used for serotyping.29 The peptide was synthesized in CDC Core Facility and purified. The peptide was solubilized in 100%, dimethyl sulphoxide, and coupled at saturating amounts (10 μg/1.5 million beads) on bead region 14 for serotyping (Table 2).
Antigen to detect recent HIV-1 infection.
Development of rIDR-M protein, which includes immuodominant region of gp41 protein from divergent subtypes, has been described before.15 The rIDR-M is an integral component of the LAg-Avidity EIA.13,15 The protein was solubilized in 100% DMSO as previously described13,15 and coupled at limiting quantities (0.04 μg/1.5 million beads) on bead region 13 for the separation of recent and long-term infections. Read outs from binding to the rIDR-M were also considered in the classification of dual (HIV-1 and HIV-2) infections.
Coupling procedure.
All regents were equilibrated at room temperature (~1 h), except the 1-ethyl-3-[3 dimethylaminopropyl] carbodiimide hydrochloride (EDC; Sigma Aldrich, St. Louis, MO), which was left at 4°C, in the dark, and in lyophilized form until needed. Bead coupling was performed as recommended by the bead manufacturer (Luminex, Austin, TX) and as described in the xMAP Cookbook with slight modifications. Briefly, each bead stock was vortexed for 20 s followed by sonication for 15 s to disperse the beads. A 120 μL aliquot of the stock microspheres, containing ~1.5 million beads, was transferred into a 1.5 mL Eppendorf tube and centrifuged at 14,000 rpm for 2 min. The supernatant was removed and the beads were washed twice with 500 μL of activation buffer (0.1 M NaH2PO4, pH 6.2; Sigma Aldrich). Each wash step included vortex and sonication of the beads followed by pelleting. Following the second wash, the beads were resuspended in 400 μL of activation buffer. Ten microliters of 50 mg/mL N-hydroxysulfosuccinimide (S-NHS, dissolved in activation buffer) (Pierce, Rockland, IL) and 10 μL of 50 μg/mL of EDC were sequentially added to the beads; mixing the beads after each addition by gently vortexing the tube. The reaction volume was adjusted to 500 μL with the addition of activation buffer and incubated at room temperature for 20 min with gentle mixing at 1,500 rpm in the dark. Mixing activates the beads by forming a semi-stable amine-reactive NHS ester. The activated beads were pelleted and washed twice with 500 μL coupling buffer [0.05 M 2-(N-morpholino) ethanesulfonic (MES) acid, pH 5]. The beads were resuspended in 200 μL of coupling buffer, vortexed, and sonicated for homogeneity. The appropriate antigen was added to the beads and the volume adjusted to 500 μL with coupling buffer followed by gentle mixing. The mixture was rotated at 1,500 rpm at room temperature in the dark for 2 h, followed by pelleting at 14,000 rpm for 2 min. The supernatant was removed and the mixture washed twice with 500 μL StabilGuard, a non-BSA blocking buffer/storage buffer (Sur-Modics, Inc., Eden Prairie, MN) and resuspended in 1 mL of StabilGuard buffer. The beads were then counted on a Luna Automatic Cell Counter (Vita Scientific, College Park, MD) and stored at 4°C protected from light until when needed. In all, three different bead regions (12, 13, and 14) were coupled with three different antigens (p24-gp41, rIDR-M, HIV-2 IDR, respectively). A scale up of coupling was performed by proportionally increasing the amounts of antigen and the beads following the scale up recommendations by the manufacturer in the Luminex xMAP Cookbook.
Multiplex assay
Coupled beads were vortexed and sonicated to ensure bead dispersal and homogeneity. A set number of beads from each bead region was prepared and used in either the monoplex format or in the multiplex format. In the monoplex format (using panels 1, 2, and 3), 1,000 beads in 50 μL volume were transferred into each well in a corning 96-well round bottom polystyrene plate (Corning Life Science, Union City, CA), while 3,000 beads (in 50 μL volume) from three bead regions were transferred into each well in a multiplex format (applies to panels 4 and 5 only). Plasma samples were diluted 1:50 in assay buffer (0.02 M Na2HPO4, 0.0036 M KH2PO4 0.5 M NaCl, 0.0054 M KCl, 0.1% Tween 20, 1% BSA, pH 7.4) and added onto the wells containing the beads. The plate was sealed with adhesive tape and the mixture was incubated for 1 h at room temperature away from light, while agitating on a multi microplate genie (Scientific Industries, Inc., Bohemia, NY). After 1 h, the plate was washed four times with wash buffer (0.02 M Na2HPO4, 0.0036 M KH2PO4 0.274 M NaCl, 0.0054 M KCl, 0.1% Tween 20, pH 7.4), rotating the plate after the second wash. The beads were resuspended in 100 μL of assay buffer containing 2 μg/mL of phycoerythrin-conjugated goat anti-human antibody (Southern Biotech, Birmingham, AL) and incubated for 30 min at room temperature while agitating in the dark. Following incubation, the beads were washed with wash buffer and resuspended in 100 μL of wash buffer. Data were acquired using the Luminex MagPix instrument (Luminex) according to the manufacturer’s instructions with the background of each bead region determined from wells with no samples added. Briefly, the instrument was programmed to read the appropriate bead regions and to acquire a minimum of 50 events in an acquisition volume of 50 μL. Signal output was measured in MFI units, which is directly proportional to the number of antibodies bound. The classification of the specimen was based on the magnitude of the MFI.
LAg-Avidity EIA
LAg-Avidity EIA was performed on all samples with HIV-1 and HIV-1/2 dual infections as described previously.13,27
Results
Monoplex optimization of diagnosis, serotyping, and recency identification
A broad range of p24-gp41 antigen concentrations were coupled on bead region 12 as illustrated in Table 2. Following a systematic optimization of antigen concentration, sample dilution, incubation times, detection antibody, assay buffer, wash conditions, and others on panel 1 specimens, the conditions clearly demonstrated that 1 μg of p24-gp41 accurately separated HIV-1 positive specimens from HIV-1 negative specimens and had the highest signal to noise ratio (data not shown) and optimal separation visually. Quantitative MFI values were used as the read out to determine the presence of HIV antibodies and the separation between negative samples (NS), and positive samples (PS).
NS had very low MFI values. However, the MFI values for the PS were several magnitudes greater than NS. Higher concentrations of p24-gp41 did not result in further increases in MFI.
The HIV-2 IDR peptide for serotyping was coupled on bead region 14 and followed similar systematic optimizations as the p24-gp41 antigen, but using panel 2 specimens. Similar trends were observed using the HIV-2 IDR peptide at an optimal concentration of 10 μg/1.5 million beads. At this concentration, there was accurate separation of HIV-2 positive specimens from HIV-2 negative specimens using panel 2.
A similar optimization procedure was performed with the rIDR-M protein, but by applying the principle of limiting antigen avidity measurement to separate recent from long-term infections. Using known specimens with recent (n = 8) and long-term infections (n = 7) (panel 3), we titrated the rIDR-M antigens for the amount with low MFI values for recent infection and high MFI for long established infections. Based on this systematic titration we experimentally determined that 0.04 μg/1.5 million beads efficiently separated recent from long-term infections.
Small panel evaluation of the multiplex assay
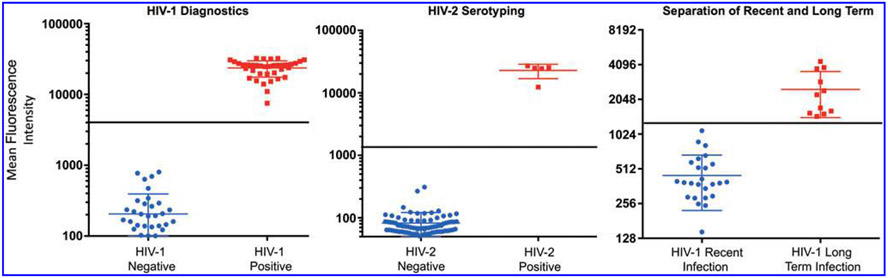
Following the initial optimization with panels 1, 2, and 3, beads were then combined in the multiplex format and blindly evaluated using panel 4 specimens. Results obtained from the multiplex assay were compared to reference results. Mean MFI of the diagnostic antigens on 45 negative specimens was ~211 (range 30–803 MFI U) (Fig. 2, left). On the other hand, HIV antibody-positive specimens had higher MFI values with a mean MFI of 23,710 (range from 7,530 MFI U to 32,395 MFI U). Based on these data and other considerations, a tentative cutoff of 4,000 MFI U was determined to maximize the sensitivity of the multiplex assay for detecting HIV-1 and dual antibody positive specimens without severely compromising on the specificity of the assay. With this tentative cutoff, the diagnostic antigen within the multiplex assay performed very well with 100% sensitivity and specificity on the 85 member panel (Fig. 2, left).
FIG. 2.
A scatter plot showing multiplex results for panel 4 (N = 85) demonstrating effective separation of specimens and distribution of MFI using beads coated with p24-gp41 (left), HIV-2 IDR (middle), and rIDR-M (right). The horizontal lines (black) show the tentative cutoffs that separate the two groups within each parameter and the middle horizontal line within each group indicates the mean value for the group. N = 85. MFI, mean fluorescent intensity.
The HIV-2 IDR peptide was designed to specifically bind to HIV-2 antibodies. Based on the data for HIV-2 IDR binding and using a similar analysis to HIV-1, a cutoff of 2,000 MFI U was determined to separate between HIV-2 positive and HIV-2 NS with maximum sensitivity and specificity. HIV-2 IDR properly classified all five HIV-2 antibody-containing specimens in the panel with 100% sensitivity and specificity (Fig. 2, middle), two of which were dual specimens (HIV-1/2).
The rIDR-M protein also performed very well in the multiplex format with panel 4 specimens. Twenty-six of the 40 HIV antibody-positive specimens in panel 4 have been characterized by the LAg-Avidity EIA as recent HIV infections. The remainder, 14 specimens were either long-term or HIV-2 specimens (Table 1). By analyzing the MFI values of the known recent samples, a cutoff of 1,250 MFI U was determined to separate recent from long-term infections. Long established infections had MFI values greater than 1,250, while recent infections had MFI values less than 1,250. Using this cutoff, the multiplex assay accurately classified all the 26 specimens as recent infections and 11 of the 14 as long-term infections (Fig. 2, right). The remainder three specimens were determined by the HIV-2 IDR to contain only HIV-2-specific antibodies. Therefore, using the 85-member panel with a cutoff of 1,250 MFI U, the multiplex assay was 100% concordant with the LAg-Avidity EIA (Fig. 2, right).
Large panel evaluation of the multiplex assay
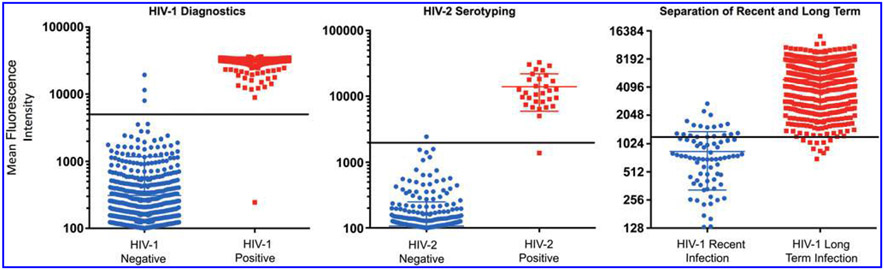
We subsequently used the settings determined using panel 4 and applied them to panel 5 specimens (n = 1,500) for larger scale evaluation of the multiplex assay. The results are presented in Figure 3, and summarized in Table 3. For the diagnostic component, 569 of 570 HIV-1 specimens were confirmed as HIV positive; three specimens tested FP in the multiplex assay by binding to p24-gp41, while there was one false negative (FN). Of the HIV-2 specimens (n = 31) there was one FN and one FP. It surmises that the sensitivity and specificity of the diagnostic arm of the multiplex assay on the 1,500 panel were 99.8% and 99.7%, respectively, while the sensitivity and specificity of serotyping were 96.8% and 99.9%, respectively. Dual infections were determined by analyzing antibody binding on all antigens. Overall, there was a 99.7% agreement between the multiplex assay and the combined reference data (EIA + Western blot + Multispot algorithm).
FIG. 3.
A scatter plot showing the distribution and antibody reactivity levels of p24-gp41 (left), HIV-2 IDR (middle), and rIDR-M (right) for Panel 5 (N = 1,500). The horizontal lines (black) show the tentative cutoffs that separate the two groups within each parameter and the middle horizontal line within each group indicates the mean value for the group. N = 1,500.
Table 3.
Comparing the Performance of the Diagnostic and Serotyping Components of the Multiplex Assay with Results from the Standard Algorithm
| Multiplex↓ | Total | Sensitivity | Specificity | |||
|---|---|---|---|---|---|---|
| HIV-1 | HIV-2 | Negative | ||||
| HIV-1 | 569 | 0 | 3 | 572 | 99.8 | 99.7 |
| HIV-2 | 0 | 30 | 1 | 31 | 96.8 | 99.9 |
| Negative | 1 | 1 | 895 | 897 | ||
| Total | 570 | 31 | 899 | 1,500 | ||
Reference results determined by EIA/Western blot/Multispot.
Shaded numbers are those with agreement between multplex classification and reference results.
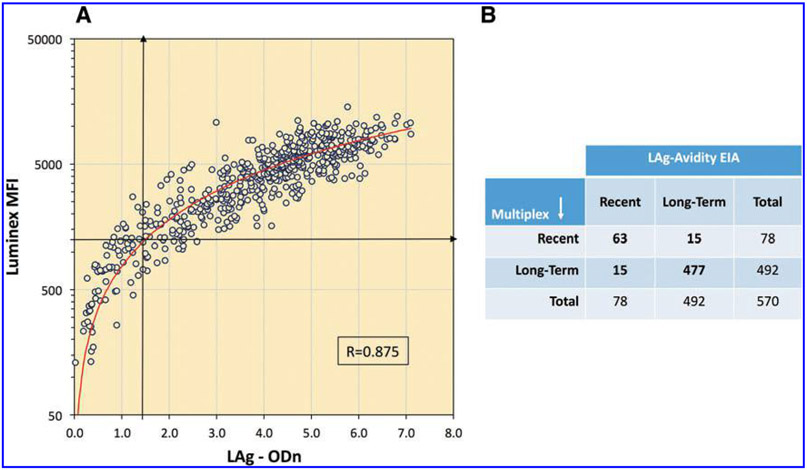
There was a high level of correlation between the LAg Avidity EIA and the multiplex assay (Spearman Rank Correlation [Rho] = 0.875, Fig. 4A) with strong overall agreement (>80%) between the two assays to distinguish recent and long-term infections. Both assays classified 78 individuals as recent with 63 being common for both assays (Fig. 4B). Additional 15 specimens that changed classification were close to the cutoff; therefore this change was as expected.
FIG. 4.
(A) Scatter plot showing correlation between LAg-Avidity EIA and multiplex assay to distinguish recent and long-term infection with red line showing polynomial best fit. Vertical arrow represents LAg cutoff of 1.5 and horizontal line represents Luminex cutoff of 1,250 MFI. (B) A 2 × 2 table summarizing recent and long-term classification by both assays. Data from all confirmed HIV-1 infections (n = 570) that include three dual infections are analyzed.
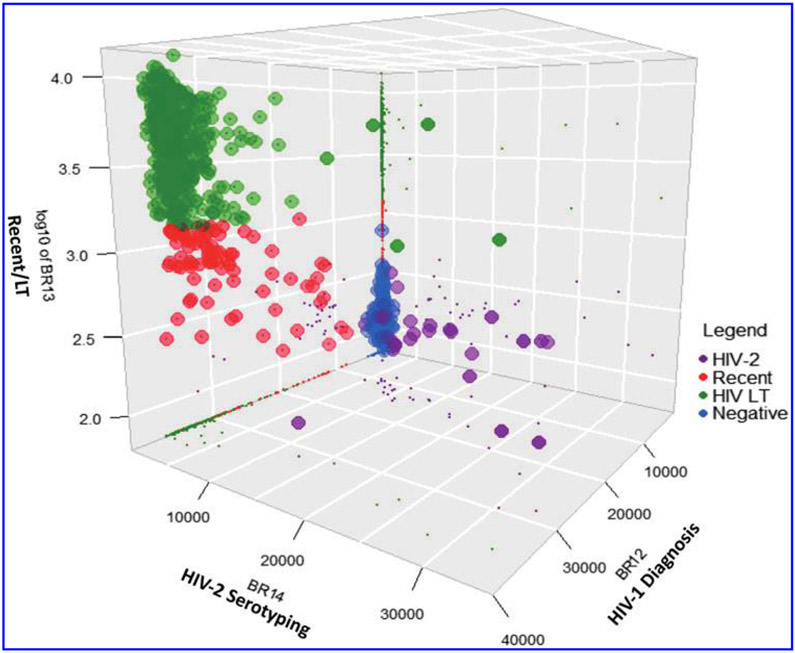
A combination of diagnostics, serotyping, and recency detection, and separations of long-term infections for panel 5 is shown as a 3D representation in Figure 5, with the different colors showing distribution of each specimen type. Note that only specimens that contained HIV-1 antibodies (HIV-1 and dual infections) were used to classify recency and long-term infections.
FIG. 5.
Three-dimensional analysis of the multiplex data for panel 5 (n = 1,500) showing partitioning of different analytes using cutoffs of 4,000 MFI U for HIV-1 positive and negative; 2,000 MFI U for HIV-2 positive and negative specimens; and 1,250 MFI U for recent and long-term infections. Three-dimensional plot with shadows on the wall (small dots) allow for proper visualization of the separations between the different parameters. The z-axis MFI is log transformed.
Multiplex assay reproducibility
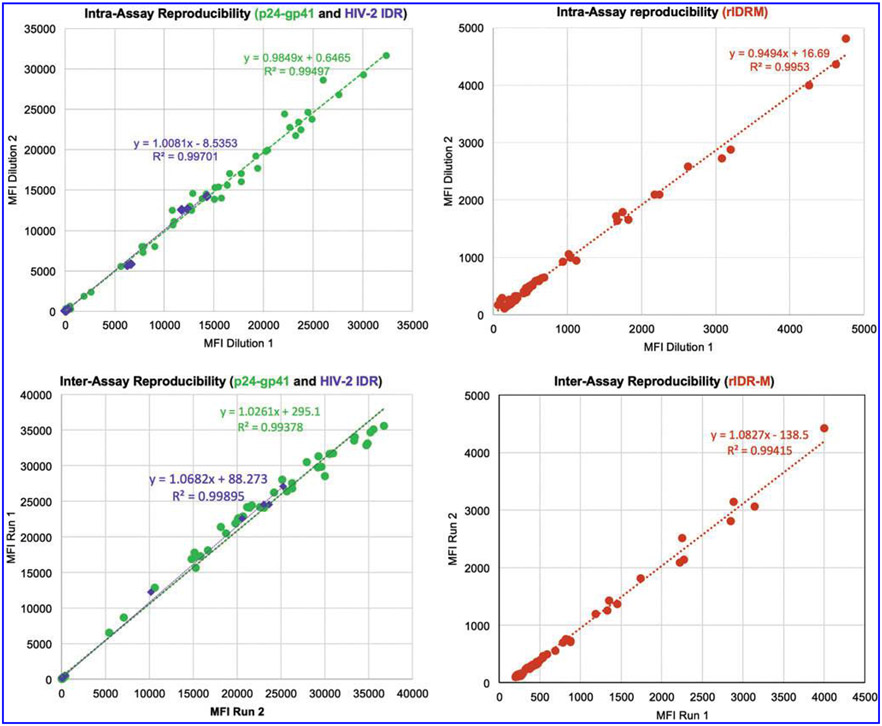
Intra- and inter-assay reproducibility of the multiplex assay were performed using 10 plasma specimens and subsequently with a subset of panel 4 specimens (n = 85) covering the dynamic range of the assay. Intra-assay reproducibility was determined using triplicate samples, while inter-assay variability was computed from at least three different runs of the same samples. The results of all three antigens in the multiplex format consistently showed high reproducibility with R2 close to 1 (Fig. 6), slope of best fit line close to 1.0, and low % coefficient of variation (%CV) (<10% in the dynamic range). For the separation of recent and long term using limiting antigen concentrations, the %CV of multiple runs was in the range of 10%–20% compared to %CVs for both the diagnostic and serotyping components of the multiplex assay (<10%).
FIG. 6.
Assay reproducibility shown as correlation in MFI. Shown is the concordance between MFI values obtained from analysis of a mixture of negative and positive specimens from panels 4 and 5. Note that specimens used for inter-assay reproducibility and intra-assay reproducibility are not necessarily the same. Line of best-fit is represented by the dotted lines with statistics shown. Intra-assay reproducibility is from three different dilutions with two dilutions compared. Inter-assay data are from two independent runs on different days. Green, purple, and brown represent single well consistencies of p24-gp41, HIV-2 IDR, and rIDR-M, respectively.
Discussion
HIV-1 prevalence and incidence are critical components of a survey to estimate the burden of HIV infections and the rate of new infections, respectively. Both of these parameters are affected by accuracy of testing algorithm. Suboptimal testing practices (e.g., use of only EIA-based algorithm) resulting in some FPs in the survey can significantly elevate HIV prevalence and incidence when using laboratory-based incidence assays.30 Further complexity comes from presence of HIV-2 in the population, especially in West Africa, which should be identified and diagnosed correctly before testing for recent HIV-1 infections. All these require multiple steps adding cost and complexity before final outcomes of the survey.
We demonstrate here the development of a multiplex assay that combines critical elements of biomarker detection to improve accuracy of HIV-1 and HIV-2 prevalence and HIV-1 incidence. The diagnostic component of the multiplex assay was performed with superior sensitivity and specificity (99.8% and 99.7%, respectively); determined by comparing results from the multiplex assay with those from reference results generated using several individual assays. The p24-gp41 antigen is a multi-epitope antigen that expresses epitopes for both capsid (p24) and envelope (gp41) antibodies, leading to increased accuracy and sensitivity. Performance of the multiplex assay was either similar or better than those of third generation EIA. However, the results show that a few samples were misclassified: 3 HIV-1 FPs, 1 HIV-2 FP, 1 HIV-1 FN, and 1 HIV-2 FN. The misclassification of FP could arise from the presence of heterophilic antibodies that cause nonspecific antibody interactions that could result in high MFI values, or could be very recent infections that were not detected by EIA/Western blot. Note that by using results from the EIA/Western blot/Multispot to compare the performance and evaluation of the multiplex assay, we assume that the sensitivity and specificity of the combined assays are 100%, which is not necessarily true. The new luminex multiplexed assay is a near-solution phase reaction kinetic assay whose rate of specific binding is increased and nonspecific binding reduced by shaking during incubations and stringent buffer system for dilutions and washes.
Another important component of the multiplex assay is its ability to serologically identify HIV-2 infections. By using a gp36 immunodominant peptide for differential typing of HIV-2, we ensured that the HIV-2 antibodies were uniquely picked up by the peptide with 96.8% and 100% sensitivity and specificity respectively. This will allow use of this multiplex assay in geographic locations where HIV-2 is endemic since it is critical to identify HIV-2 before HIV-1 incidence can be estimated. As described earlier, the rIDR-M antigen is designed to bind to HIV-1 antibodies and is nonreactive to HIV-2 antibodies.15 Therefore, in the absence of HIV-2 IDR peptide, HIV-2 specimens would be misclassified as recent infections. The multiplex assay can therefore reduce the misclassification rate that could possibly result from the presence of HIV-2 antibodies, while providing information about prevalence of HIV-2 in the survey.
We previously described development of a rapid incidence-prevalence test,14 which can simultaneously diagnose HIV infection and distinguish recent from long-term infection. However, this test will require HIV-2-specific serotyping, especially in areas where HIV-2 is prevalent, to confirm HIV-1 recent infections. To our knowledge, this is the first multiplex assay with this triad capability for HIV. Due to multiplex capability, this test can eliminate multiple layers of testing that is traditionally required in a survey with the aim to determine HIV prevalence and incidence. Magpix platform is robust and easy to use in any centralized laboratory capable of performing EIAs and similar serologic assays.
Comparative data with LAg-Avidity EIA demonstrate that the assay show high correlation (R = 0.84) with LAg-EIA (Fig. 4). These data allowed us to infer cutoff and the mean duration of recent infection (MDRI) from this correlation with LAg EIA. Testing of longitudinal specimens collected from incident infections is in progress to finalize MDRI determination. Similarly, determination of the proportion of false recent (PFR) requiring testing of large number of specimens with long-term infections is also in progress. As with most antibody-based assays, including LAg-EIA, we expect some level of misclassification among individuals with suppressed viral load, which may include elite controllers and persons on ART. Therefore, addition of viral load (VL) may remain as an important second step of recent infection testing algorithm.31 Testing of specimens from elite controllers and persons of ART will follow in the future to better quantify misclassification and mitigation by VL testing.
Previously, a multiplex assay has been described for the estimation of recent infection using a multi-analyte approach for recency, in an attempt to reduce PFR and increase the accuracy of incidence estimates.12,32,33 However, this assay did not incorporate diagnostic and serotyping component. In addition, subtype-specific antibody bias continues to be a problem for many assays because they do not include multi-subtype antigens that are critical for recent infection detection. A decreased reactivity or antibodies of reduced avidity to antigens from different subtypes would result in misclassification. One reason the LAg-Avidity EIA has been very useful and superior in the field is its ability to reduce subtype-specific bias in the population, a feature accorded by the multi-subtype gp41 recombinant protein, rIDR-M, and the ability to measure antibody avidity in a single well.13,17 Our newly developed multiplex assay captures these properties of the LAg-Avidity EIA as it is developed using the same principle and the same antigen. The diversity of the panel as described suggests that the multiplex assay might perform similarly across diverse subtypes with minimal variability. However, more data are needed to quantify this properly. When results from the LAg-Avidity EIA were compared to those from the multiplex assay using panels 4 and 5, we found that the multiplex assay at a cutoff of 1,250 MFI U correlated strongly with the LAg EIA at a cutoff of 1.5 ODn with high correlation and similar number classified as recent (Figs. 3 and 4). However, the wider dynamic range of the multiplex assay suggests that the MDRI may be increased without compromising on the accuracy of incidence estimates. A thorough analysis of the multiplex assay using a panel of longitudinal specimens will further confirm this hypothesis.
Our data also demonstrate high precision of multiplex assay (Fig. 6). In addition to analyzing for inter- and intra-assay reproducibility, we analyzed the quantitative agreement from separate couplings; inter-coupling consistency was high and the results showed low %CVs and R2 > 0.95 (data not shown). Our results also show that multiplexing did not change the absolute MFI values when compared with monoplexing, as there is a very strong correlation between monoplexing and multiplexing (data not shown). We also analyzed reproducibility by reading plates at different times after assay completion and the results show a stable signal up to 72 h after assay completion with plates stored at 4° in the dark (data not shown). There was a small reduction in signal intensity after 72 h for plates that were stored at room temperature in the dark. However, MFI signals were stable at RT for up to 8 h. Our results from various reproducibility tests show that the assay is highly reproducible within the various parameters listed as indicated by low %CV and R2 closed to 1. Although the multiplex assay shows high reproducibility we are currently exploring the possibility of further minimizing variations by using a calibrator specimen, similar to LAg-Avidity EIA.
At present, the estimation of recent and long-term infections requires a series of tests that are complex, difficult, and expensive. The increase in cost in performing different assays, on different platforms with different expertise needed may not be available in all settings. The multiplex assay is unique and presents a platform that can reduce the number of assays in the surveys. Note that other tests in the incidence algorithm, such as HIV viral load and anti-retroviral (ARV) detection that are not part of the multiplex assay will have to be performed separately. In an era of algorithm-driven diagnosis and determination of recent/long-term infections,18,34,35 it is profoundly important that a platform such as the multiplex assay exists to accommodate new biomarkers of infection or classification associated with the disease in an attempt to improve on the accuracy of incidence estimates. We are therefore exploring the possibility of adding more analytes such as the integrase antigen (p31) to the assay. Other advantages to the use of the multiplex assay include its simplicity, minimal sample volume requirement, and low cost. Being quantitative, the multiplex assay is independent of subjective bias and uses reagents that are easily made in the lab. This study suggests that the multiplex assay has strong potential to revolutionize incidence surveillance. However, a thorough analysis of more data (especially longitudinal specimens and long-term specimens) is required to define important parameters such as MDRI, the proportion misclassified as false recent (PFR) among persons with long-term (>1 year) HIV-1 infections, subtype-specific performance, and misclassification among AIDS patients with and without the use of antiretroviral drugs. We also recognize potential impact of pre-exposure prophylaxis on MDRI and PFR and need to include appropriate specimens for further evaluations. These analyses will be followed by a field validation of the assay comparing it with the standard LAg-Avidity EIA.
In summary, our study presents a multiplexed system demonstrating that diagnosis, serotyping, and recent infection determination of HIV can be simultaneously performed in a single test, using very limited sample volume (Fig. 1). Our results show that the assay is robust and precise with consistently low inter- and intra-assay variability. As described, the system weeds off some of the key contributing and confounding sources of error in the estimation of recent HIV-1 infection while providing data for HIV-1 and HIV-2 prevalence. Simultaneous classification of individuals as HIV positive or negative, if positive, HIV-1 or HIV-2, and if HIV-1, recent or long term, would have a major impact in public health surveillance as we strive to control the HIV epidemic.
Acknowledgments
This project has been supported in part by the President’s Emergency Plan for AIDS Relief (PEPFAR) through the Centers for Disease Control and Prevention (CDC), Atlanta. E.L.Y. was supported by the ASM/CDC program in infectious disease and public health microbiology postdoctoral research fellowship. We thank Meade Morgan for statistical assistance, help with the 3D plot, and critical review of the article.
Footnotes
Disclaimer
The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or other funding agencies.
Author Disclosure Statement
As an inventor of LAg-Avidity EIA, which is available as a commercial kit, B.S.P. receives a portion of the royalties from the sale of the LAg-Avidity EIA as per policy of the U.S. Government. E.L.Y declares no conflicts of interest.
References
- 1.UNAIDS: AIDS Info 2015. Web Page. Available at www.aidsinfo.unaids.org, accessed May 7, 2017.
- 2.Song R, Hall HI, Green TA, Szwarcwald CL, Pantazis N: Using CD4 data to estimate HIV incidence, prevalence, and percent of undiagnosed infections in the United States. J Acquir Immune Defic Syndr 2017;74:3–9. [DOI] [PubMed] [Google Scholar]
- 3.Barin F, Meyer L, Lancar R, et al. : Development and validation of an immunoassay for identification of recent human immunodeficiency virus type 1 infections and its use on dried serum spots. J Clin Microbiol 2005;43:4441–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kothe D, Byers RH, Caudill SP, et al. : Performance characteristics of a new less sensitive HIV-1 enzyme immunoassay for use in estimating HIV seroincidence. J Acquir Immune Defic Syndr 2003;33:625–634. [DOI] [PubMed] [Google Scholar]
- 5.Parekh BS, Kennedy MS, Dobbs T, et al. : Quantitative detection of increasing HIV type 1 antibodies after seroconversion: A simple assay for detecting recent HIV infection and estimating incidence. AIDS Res Hum Retroviruses 2002;18:295–307. [DOI] [PubMed] [Google Scholar]
- 6.Parekh BS, Pau CP, Kennedy MS, Dobbs TL, McDougal JS: Assessment of antibody assays for identifying and distinguishing recent from long-term HIV type 1 infection. AIDS Res Hum Retroviruses 2001;17:137–146. [DOI] [PubMed] [Google Scholar]
- 7.Rawal BD, Degula A, Lebedeva L, et al. : Development of a new less-sensitive enzyme immunoassay for detection of early HIV-1 infection. J Acquir Immune Defic Syndr 2003;33:349–355. [DOI] [PubMed] [Google Scholar]
- 8.Suligoi B, Galli C, Massi M, et al. : Precision and accuracy of a procedure for detecting recent human immunodeficiency virus infections by calculating the antibody avidity index by an automated immunoassay-based method. J Clin Microbiol 2002;40:4015–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suligoi B, Massi M, Galli C, et al. : Identifying recent HIV infections using the avidity index and an automated enzyme immunoassay. J Acquir Immune Defic Syndr 2003;32:424–428. [DOI] [PubMed] [Google Scholar]
- 10.Janssen RS, Satten GA, Stramer SL, et al. : New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. JAMA 1998;280:42–48. [DOI] [PubMed] [Google Scholar]
- 11.Andersson E, Shao W, Bontell I, et al. : Evaluation of sequence ambiguities of the HIV-1 pol gene as a method to identify recent HIV-1 infection in transmitted drug resistance surveys. Infect Genet Evol 2013;18:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtis KA, Kennedy MS, Charurat M, et al. : Development and characterization of a bead-based, multiplex assay for estimation of recent HIV type 1 infection. AIDS Res Hum Retroviruses 2012;28:188–197. [DOI] [PubMed] [Google Scholar]
- 13.Duong YT, Qiu M, De AK, et al. : Detection of recent HIV-1 infection using a new limiting-antigen avidity assay: Potential for HIV-1 incidence estimates and avidity maturation studies. PLoS One 2012;7:e33328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granade TC, Nguyen S, Kuehl DS, Parekh BS: Development of a novel rapid HIV test for simultaneous detection of recent or long-term HIV type 1 infection using a single testing device. AIDS Res Hum Retroviruses 2013;29:61–67. [DOI] [PubMed] [Google Scholar]
- 15.Wei X, Liu X, Dobbs T, et al. : Development of two avidity-based assays to detect recent HIV type 1 seroconversion using a multisubtype gp41 recombinant protein. AIDS Res Hum Retroviruses 2010;26:61–71. [DOI] [PubMed] [Google Scholar]
- 16.Hu DJ, Vanichseni S, Mock PA, et al. : HIV type 1 incidence estimates by detection of recent infection from a cross-sectional sampling of injection drug users in Bangkok: Use of the IgG capture BED enzyme immunoassay. AIDS Res Hum Retroviruses 2003;19:727–730. [DOI] [PubMed] [Google Scholar]
- 17.Kassanjee R, Pilcher CD, Keating SM, et al. : Independent assessment of candidate HIV incidence assays on specimens in the CEPHIA repository. AIDS 2014;28:2439–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konikoff J, Brookmeyer R, Longosz AF, et al. : Performance of a limiting-antigen avidity enzyme immunoassay for cross-sectional estimation of HIV incidence in the United States. PLoS One 2013;8:e82772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nesheim S, Parekh B, Sullivan K, et al. : Temporal trends in HIV Type 1 incidence among inner-city childbearing women in Atlanta: Use of the IgG-capture BED-enzyme immunoassay. AIDS Res Hum Retroviruses 2005;21:537–544. [DOI] [PubMed] [Google Scholar]
- 20.Price MA, Wallis CL, Lakhi S, et al. : Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res Hum Retroviruses 2011;27:5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rehle T, Shisana O, Pillay V, Zuma K, Puren A, Parker W: National HIV incidence measures—New insights into the South African epidemic. S Afr Med J 2007;97:194–199. [PubMed] [Google Scholar]
- 22.Saphonn V, Parekh BS, Dobbs T, et al. : Trends of HIV-1 seroincidence among HIV-1 sentinel surveillance groups in Cambodia, 1999–2002. J Acquir Immune Defic Syndr 2005;39:587–592. [PMC free article] [PubMed] [Google Scholar]
- 23.Cooley GM, Mitja O, Goodhew B, et al. : Evaluation of multiplex-based antibody testing for use in large-scale surveillance for yaws: A Comparative Study. J Clin Microbiol 2016;54:1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fonseca BP, Marques CF, Nascimento LD, et al. : Development of a multiplex bead-based assay for detection of hepatitis C virus. Clin Vaccine Immunol 2011;18:802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smits GP, van Gageldonk PG, Schouls LM, van der Klis FR, Berbers GA: Development of a bead-based multiplex immunoassay for simultaneous quantitative detection of IgG serum antibodies against measles, mumps, rubella, and varicella-zoster virus. Clin Vaccine Immunol 2012;19:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talha SM, Saviranta P, Hattara L, et al. : Array-in-well platform-based multiplex assay for the simultaneous detection of anti-HIV- and treponemal-antibodies, and Hepatitis B surface antigen. J Immunol Methods 2016;429:21–27. [DOI] [PubMed] [Google Scholar]
- 27.Duong YT, Kassanjee R, Welte A, et al. : Recalibration of the limiting antigen avidity EIA to determine mean duration of recent infection in divergent HIV-1 subtypes. PLoS One 2015;10:e0114947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mastro TD: Determining HIV incidence in populations: Moving in the right direction. J Infect Dis 2013;207:204–206. [DOI] [PubMed] [Google Scholar]
- 29.Qiu M, Liu X, Jiang Y, et al. : Current HIV-2 diagnostic strategy overestimates HIV-2 prevalence in China. J Med Virol 2009;81:790–797. [DOI] [PubMed] [Google Scholar]
- 30.UNAIDS/WHO working group on global HIV/AIDS and STI surveillance. Monitoring the Impact of HIV Epidemic Using Population-Based Surveys, Nov 25, 2015. Available at www.who.int/hiv/pub/guidelines/si-guidelines-population-survey/en/ accessed September 14, 2018.
- 31.Kassanjee R, Pilcher CD, Busch MP, et al. : Viral load criteria and threshold optimization to improve HIV incidence assay characteristics. AIDS 2016;30:2361–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curtis KA, Hanson DL, Kennedy MS, Owen SM: Evaluation of a multiplex assay for estimation of HIV-1 incidence. PLoS One 2013;8:e64201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtis KA, Kennedy MS, Owen SM: Longitudinal analysis of HIV-1-specific antibody responses. AIDS Res Hum Retroviruses 2014;30:1099–1105. [DOI] [PubMed] [Google Scholar]
- 34.Brookmeyer R, Konikoff J, Laeyendecker O, Eshleman SH: Estimation of HIV incidence using multiple biomarkers. Am J Epidemiol 2013;177:264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brookmeyer R, Laeyendecker O, Donnell D, Eshleman SH: Cross-sectional HIV incidence estimation in HIV prevention research. J Acquir Immune Defic Syndr 2013;63 Suppl 2:S233–S239. [DOI] [PMC free article] [PubMed] [Google Scholar]