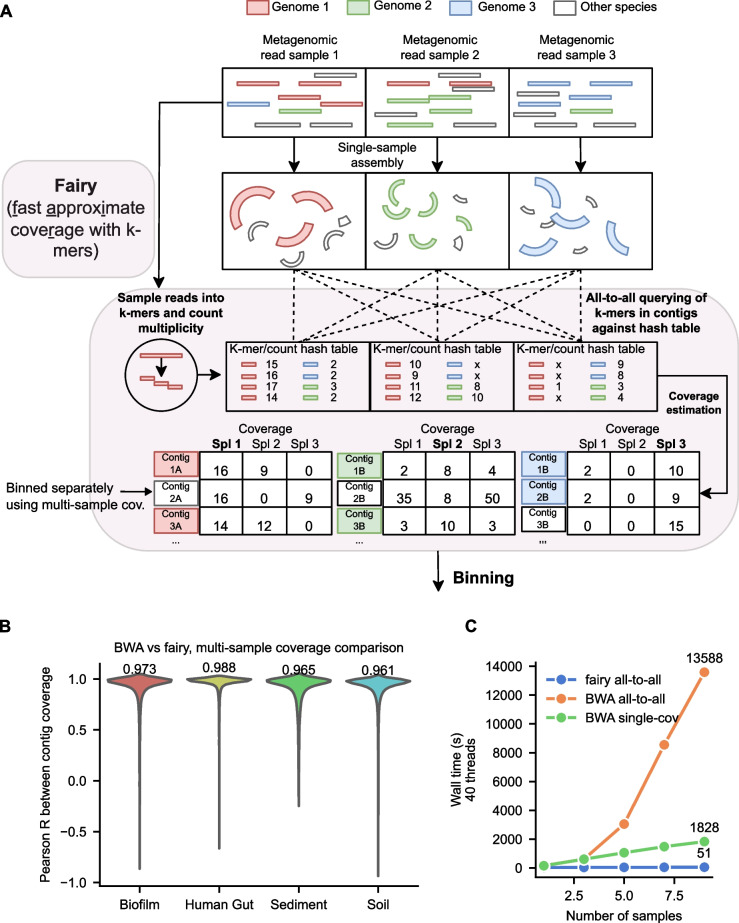
Fig. 1.
Fast approximate k-mer coverage estimates for multi-sample metagenomic binning. A Outline of fairy’s k-mer-based algorithm. Fairy’s processing steps are outlined in light red. Fairy indexes (or sketches) the reads into subsampled k-mer-to-count hash tables. K-mers from contigs are then queried against the hash tables to estimate coverage. Finally, fairy’s output is used for binning and is compatible with several binners (e.g., MetaBAT2, MaxBin2). Notice that white contigs (e.g., Contig 2A) have similar coverage to the colored contigs in the same sample, but additional samples help clarify that the white contigs should be binned separately. B Pearson R values between fairy and BWA’s multi-sample coverages for contigs in an arbitrary assembly from the dataset. Median values are shown above the plots. C Wall time with 40 threads for fairy vs BWA on 1, 3, 5, 7, and 9 gut samples in all-to-all mode (multi-sample) and single-sample mode