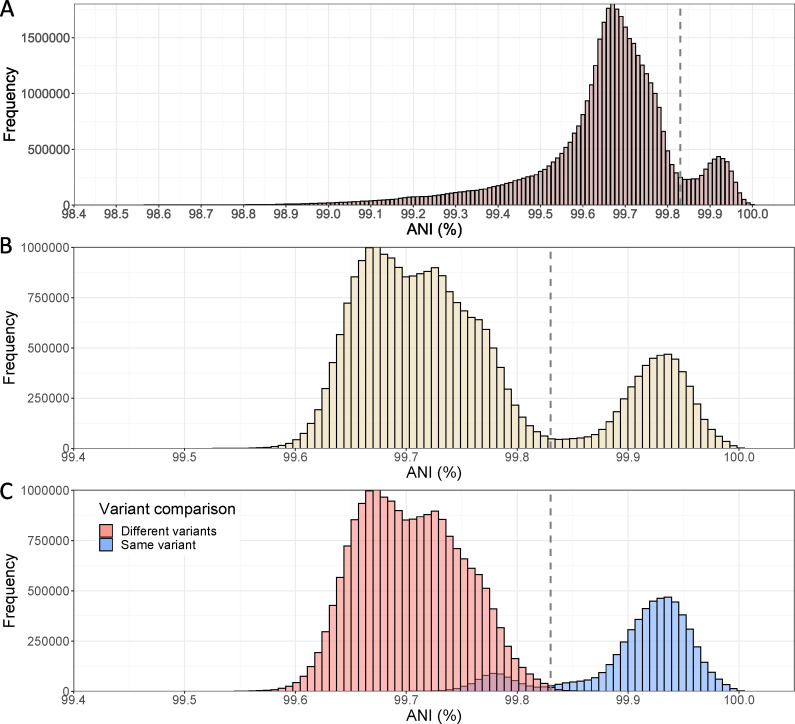
Fig 3.
ANI histograms of the SARS-CoV-2 genomes sequenced by the US CDC. (A) Histogram with a random subset of 6,481 genomes (Ns allowed) belonging to the Alpha (n = 1,250), Beta (n = 534), Delta (n = 1,250), Gamma (n = 1,250), Epsilon (n = 947), and Omicron (n = 1,250) VOCs was analyzed, that is, 41,996,880 pairs in total. A subtle ANI gap at around 99.8% can be observed (see vertical line). (B) Histogram with 5,041 high-quality genomes (i.e., without Ns) belonging to the Alpha (n = 1,250), Beta (n = 99), Delta (n = 1,250), Gamma (n = 1,035), Epsilon (n = 157), and Omicron (n = 1,250) VOCs was analyzed, that is, 27,620,280 pairs in total. After removing genomes with Ns, the data reveal a clearer bimodal distribution and a more pronounced ANI gap for the SARS-CoV-2 genomes. (C) Histogram shows the same data as in B, but the bars are colored based on whether the genomes compared are assigned to the same variants by WHO (in blue) or not (in pink). Note the limited overlap between the two groups.