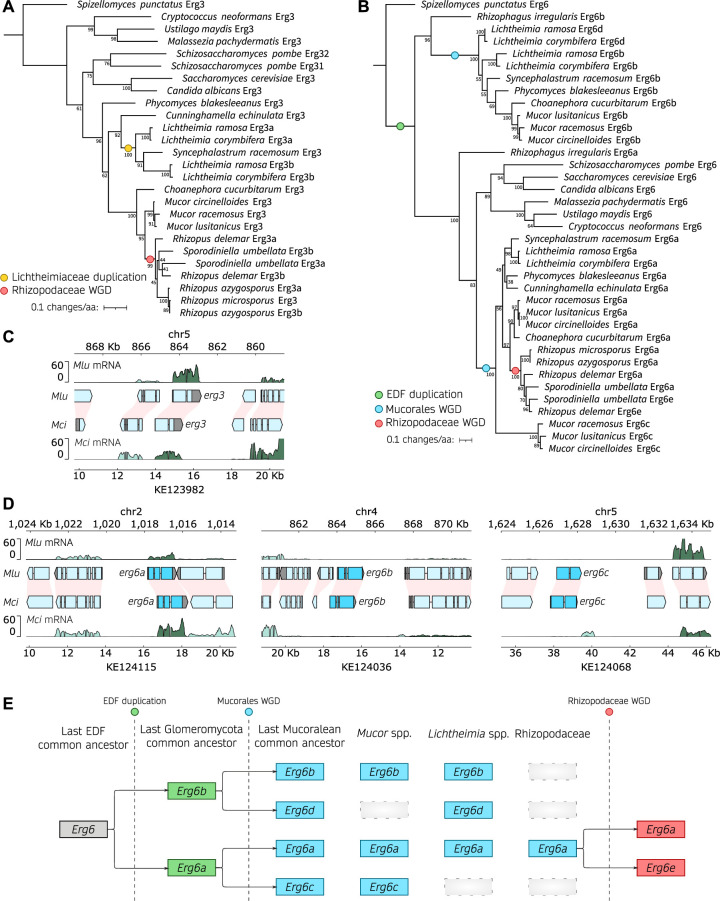
Fig 2.
Erg3 and Erg6 evolutionary history across the Mucoromycota. (A, B) Phylogenetic tree depicting the Erg3 (A) and Erg6 (B) amino acid sequence alignments in clinically relevant fungi, with a specific focus on Mucorales. The Erg protein sequences from the chytrid Spizellomyces punctatus were employed as outgroups to root the trees. Additionally, the Erg6 sequence from Rhizophagus irregularis, a member of the Glomeromycota and the closest non-Mucoromycota sequence, was included to provide clarity on duplication events in (B). Noteworthy branching events, including whole-genome duplications (WGD) or gene duplications, are denoted by color-coded circles. Node support is represented as a percentage based on 1,000 bootstrapping iterations. The branch length is scaled to 0.1 substitutions per amino acid, as indicated in the legend. (C, D) Plots illustrating gene transcription across M. lusitanicus (Mlu) and M. circinelloides (Mci) genomic regions containing erg3 (C) and erg6 orthologs (D). The genomic coordinates for both species are displayed above (Mlu) and below (Mci) each plot. Gene annotation is depicted as arrowed blocks, indicating the direction of transcription and the boundaries of exons and introns. The coloring scheme assigns cyan blue to erg coding sequences, light blue to the remaining coding sequences, and gray to untranslated regions. Interspecies synteny among genes is depicted as pink shading. Gene transcription is shown as rRNA-depleted RNA read coverage, with the forward strand represented in dark green and the reverse strand in light green, enabling assessment of sense and antisense transcription. In some instances, the x-axes were inverted, and in those cases, the forward and reverse strand orientations were also inverted to enhance visualization and clarification. (E) Model illustrating the inferred evolutionary history of Erg6 across the early-diverging fungi (EDF).