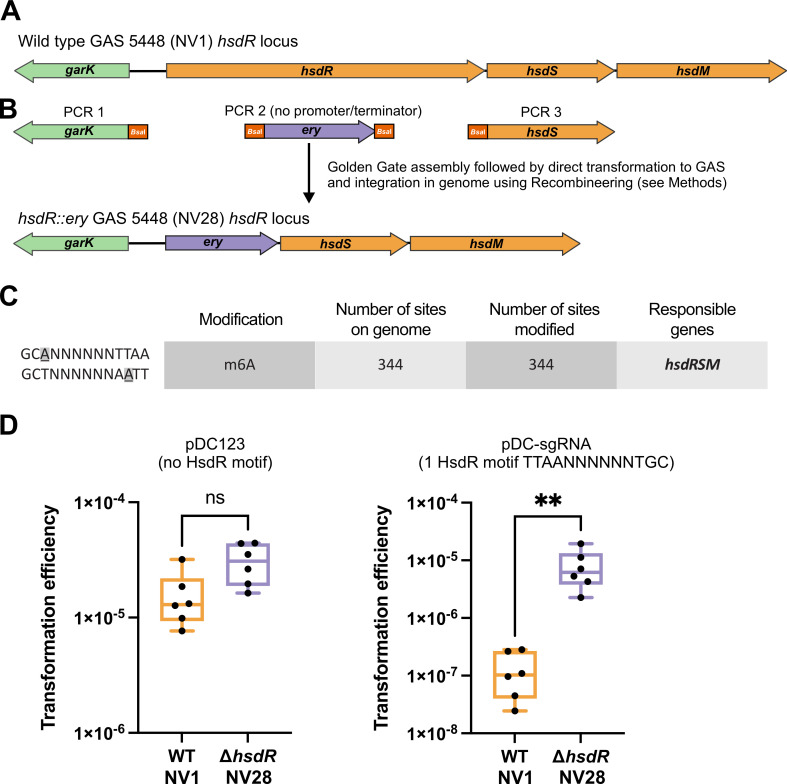
Fig 1.
Golden Gate assembly and recombineering to create a highly transformable hsdR mutant in M1T1 GAS strain 5448. (A) Schematic representation of the genomic organization of the hsdRSM locus in WT GAS strain M1 5448 (NV1). Gene annotations were made by Prokka (25). (B) Three PCRs were performed each containing unique BsaI restriction sites that allow for scarless Golden Gate assembly of a linear fragment in which hsdR is replaced by an erythromycin resistance cassette (ery). PCR 1 contains 1,392 bp upstream of hsdR (including the garK gene and excluding the start codon of hsdR), PCR 2 contains the 785 bp promoterless and terminatorless ery cassette (23) flanked by BsaI sites, while PCR 3 contains a 1,238 bp downstream region of hsdR (including its stop codon and hsdS). After Golden Gate assembly with BsaI and T4 ligase (see Materials and Methods), the resulting 3,383-bp ligation product was purified and directly used to transform electrocompetent NV2 (NV1 + pAV488) cells that were grown with 1 mM of β-D-1-thiogalactopyranoside to induce the recombineering system present on plasmid pAV488 resulting in strain NV3 (NV2, hsdR::ery). Recombineering of GAS using pAV488 is described in more detail elsewhere (Andrew Varble, unpublished data). Strain NV3 was plasmid cured resulting in strain NV28 (NV1, hsdR::ery). The genomic organization of the hsdRSM locus in strain NV28 (hsdR::ery) is shown. (C) Methylation motifs identified in strains NV1, NV28, and NV6 using SMRT sequencing (see Materials and Methods for details). (D) Transformation efficiencies of WT GAS 5448 (NV1) and the hsdR::ery mutant (NV28) are shown. Left: strains were transformed with 140 ng (~50 fmol) of plasmid pDC123 (24) that does not contain an HsdR-motif. Right: strains were transformed with 247 ng (~95 fmol) of plasmid pDC-sgRNA (see Fig. 2C) that contains a single HsdR-motif. Each dot represents a replicate, and a Kolmogorov-Smirnov test was used to calculate statistically significant differences between the two strains (**P value < 0.005).