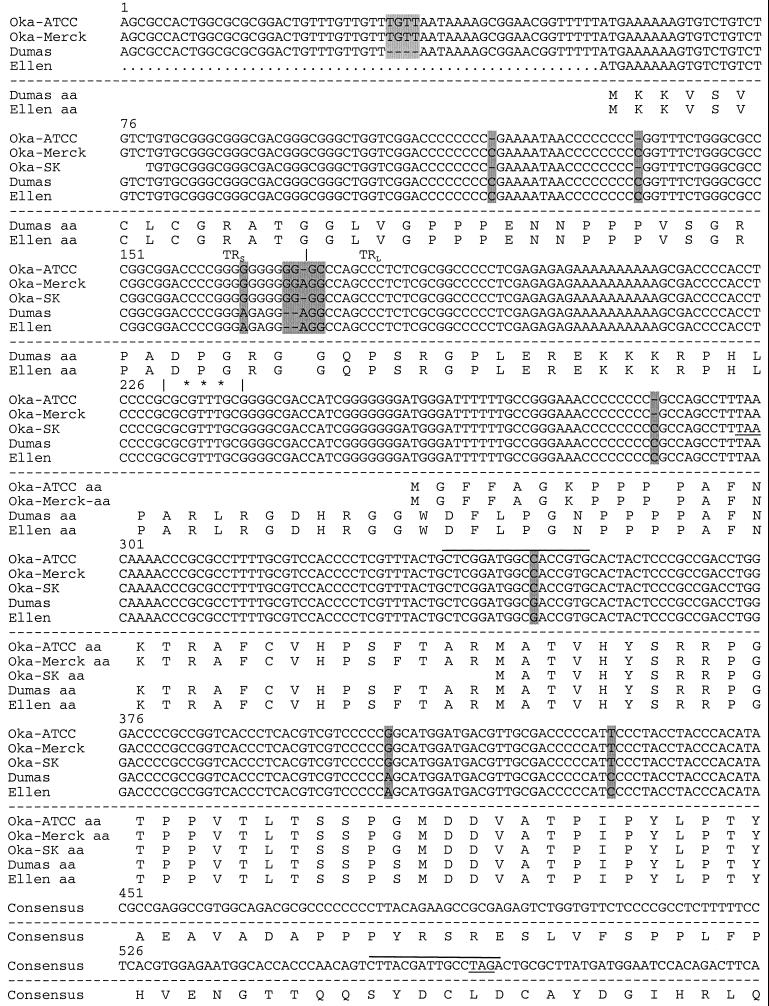
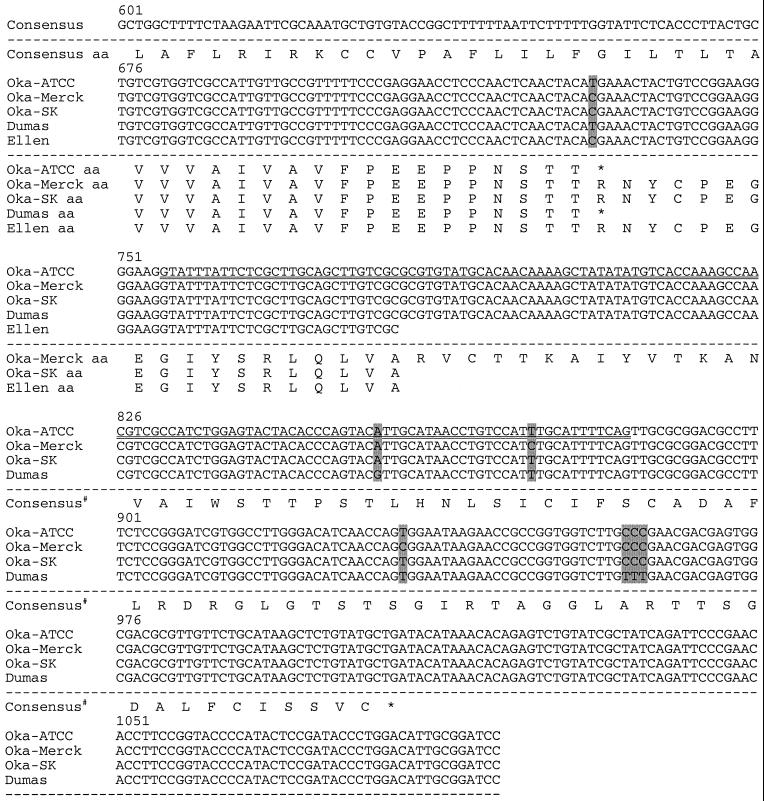
FIG. 2.
Sequence comparison of VZV genomes near ORF S/L. The nucleotide sequences of Oka-ATCC, Oka-Merck, Oka-SK, Dumas, and Ellen as displayed all begin within TRS; the TRS/TRL boundary is identified with a vertical line, and nucleotide deletions (−) are indicated. Sequence differences between the strains are shaded. The predicted amino acid sequences are displayed beneath the nucleotide sequences. Consensus# represents the Oka-Merck and Oka-SK consensus amino acids. Positions of the transcription initiation site (∗∗∗), the 130-nt intron (⩵⩵), potential termination sites (underline), and primer sites used to generate anti-ORF S/L serum (overline) are indicated. The terminator at position 567 would truncate the S/L protein encoded by Oka-Merck if it were to initiate at the same ATG as Dumas.