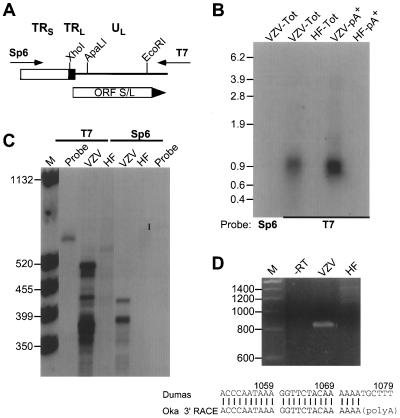
FIG. 3.
Mapping the ORF S/L mRNA. (A) Depiction of pGS/LC3, a plasmid clone of a PCR product spanning the TRL/TRS junction of Oka-ATCC. Directions of the T7 and Sp6 transcripts generated from this insert are indicated; the Sp6-transcribed RNA would be the same sense as ORF S/L mRNA. (B) Northern analysis of RNA from ORF S/L. Ten micrograms of whole-cell (Tot) or 1 μg of poly(A)+ (pA+) RNA isolated from uninfected (HF) or VZV-infected (VZV) fibroblasts was subjected to Northern analysis with either the T7/XhoI (T7) or Sp6/EcoRI (Sp6) probe. The autoradiographic image of this analysis is shown, with sizes of the RNA markers indicated in kilobases. (C) RNase protection analysis of ORF S/L RNA. The T7/XhoI (T7) in vitro-transcribed probe was hybridized to whole-cell RNA isolated from VZV-infected (VZV) or uninfected (HF) fibroblasts and digested with RNase ONE for 60 min. The products were separated by electrophoresis in a denaturing polyacrylamide gel, and the gel was exposed to X-ray film. Positions and sizes (in nucleotides) of the molecular weight standards (lane M) are indicated at the left. The closed circle adjacent to the lane marked probe identifies the position of unhybridized probe. (D) 3′ RACE analysis of ORF S/L RNA. Whole-cell RNAs from uninfected (HF) and infected (VZV) cells were reverse transcribed and subjected to RT-PCR. The products of these reactions and a control reaction that was done without reverse transcriptase (−RT) were electrophoresed on a 4% 3:1 agarose gel and subsequently stained with ethidium bromide. The RT-PCR product shown in the VZV lane was directly sequenced. The junction of the Oka ORF S/L mRNA and the poly(A) region are depicted and aligned with the Dumas sequence (accession no. X04370). The numbers refer to Dumas genomic DNA. Sizes of the DNA markers (lane M) are shown in nucleotides at the left. Autoradiograms and negatives of stained gels were scanned with a GS-250 imaging densitometer (Bio-Rad, Hercules, Calif.). Photographic quality images were generated using Molecular Analyst (Bio-Rad) and Adobe Photoshop (Adobe Systems, Mountain View, Calif.) software.