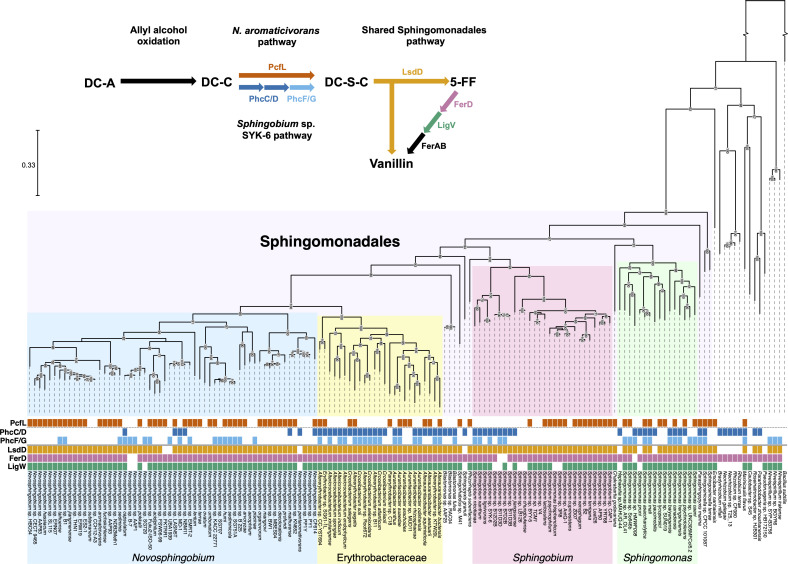
Fig 10.
Order Sphingomonadales contains two pathways for conversion of DC-C to DC-S-C and a conserved pathway for DC-S-C catabolism. Phylogeny constructed based on the bacterial reference genes of Alphaproteobacteria containing homologs (>50% amino acid identity, >70% query coverage) of at least two enzymes found in the β-5 linked aromatic catabolic pathways characterized in N. aromaticivorans or Sphingobium sp. SYK-6. Homologs found in each species are marked by colored boxes. Clades are labeled and color-coded. The scale bar indicates the number of nucleotide substitutions per sequence site. The gap in the outgroup corresponds to 1.5 on the scale bar. A simplified diagram of the DC-A catabolic pathways in N. aromaticivorans and Sphingobium sp. SYK-6 is shown.