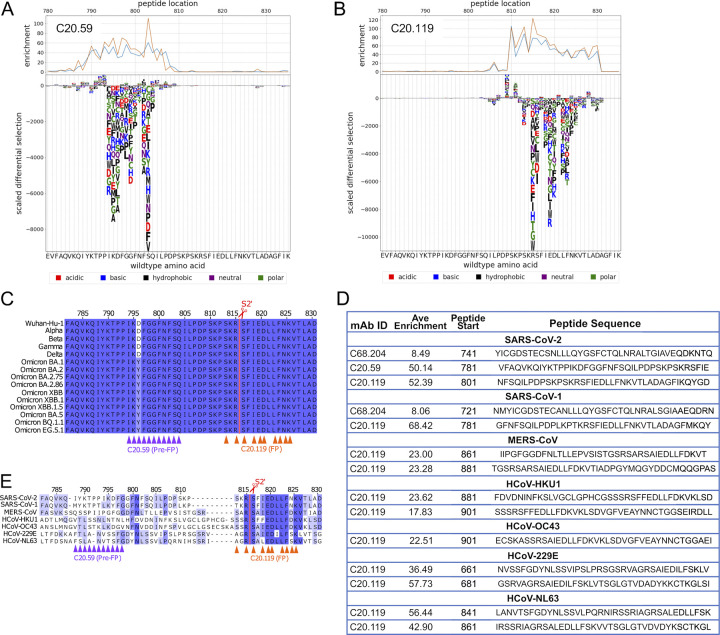
Fig 6. Epitope mapping by PhIP-seq identified linear epitopes in and around the FP region of S2 subdomain for mAbs C20.59, C20,119, and C68.204.
(A,B) Each mAb was incubated with the phage-display spike peptide library, which consisted of 31 amino acid (aa) peptides with a 30 aa overlap (Phage-DMS library). Along with the Wuhan-Hu-1 wildtype sequences, each peptide was also represented with the central residue mutated to every possible amino acid to observe loss of mAb binding, or escape, with each mutation. The top graphs display the binding enrichment of peptides in that region in the WH-1 library for each mAb, with experimental replicates shown in the orange and blue lines for C20.59 (A) and C20.119 (B). The bottom logo plots depict the effect of single mutations on binding of the mAb to the peptide. The height of the letter corresponds to the size of the effect that mutation had on the binding or the scaled differential enrichment with the mutation compared to the wildtype sequence. Mutations that resulted in a positive (>0) scaled differential enrichment are changes that increased mAb binding to the peptide, whereas a negative scaled enrichment is due to reduced binding with that mutation. The letter color signifies the chemical property of the amino acid. The WH-1 or wildtype sequence is shown on the x axis. (C,D) Sequence alignments across SARS-CoV-2 VOCs (C) or HCoVs (E) of the region upstream of and including FP (WH-1 aa 782–830). The key escape residues in the epitopes for C20.59 (purple) and C20.119 (orange) are noted under the alignment by the triangles. The color of the residues in the alignment denotes the percent identify at that site across the sequences shown. The S2’ cleavage is marked by the red line and scissors. (D) Peptides enriched in the pan-CoV library after binding with the three S2 mAbs C20.59, C20.119, and C68.204. The pan-CoV library included peptides covering the entire spike sequence from HCoVs SARS-CoV-2 WH-1, SARS-CoV-1, MERS-CoV, HCoV-HKU1, HCoV-OC43, HCoV-229E, and HCoV-NL63. Each mAb was run as technical replicates and the enrichment scores were averaged. Significantly enriched peptides (p<0.5) were identified and those peptides with an average enrichment greater than the off-target S1 regions are shown for C20.59, C20.119, and C68.204.