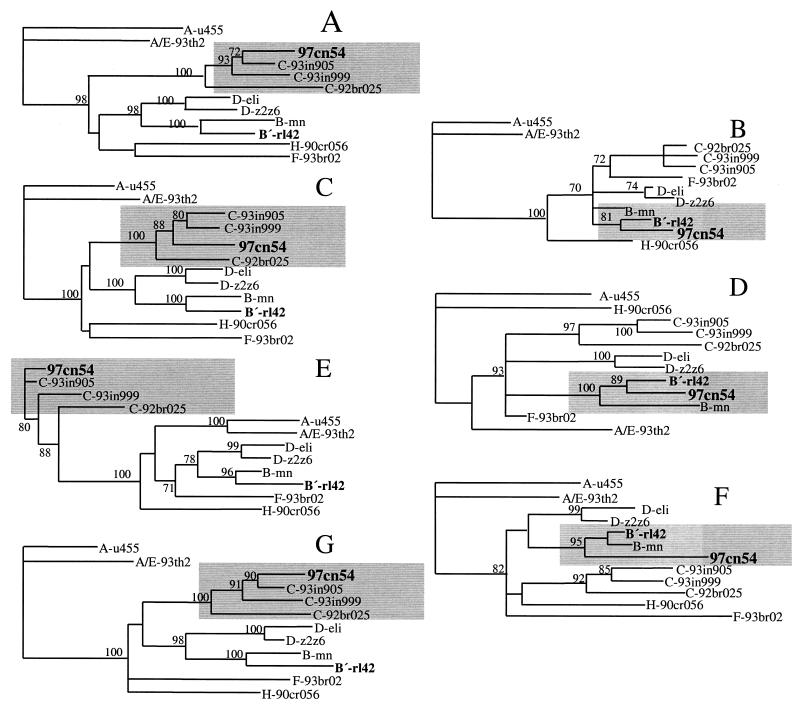
FIG. 3.
Phylogenetic relationship of different regions within the 97cn54-derived gag-pol reading frames to standard representatives of the major HIV-1 (group M) subtypes. Phylogenetic trees were constructed using the neighbor-joining method based on the following sequence stretches: nucleotides 1 to 478 (A), 479 to 620 (B), 621 to 1290 (C), 1291 to 1830 (D), 1831 to 2220 (E), 2221 to 2520 (F), and 2521 to 2971 (G). The indicated positions refer to the first nucleotide of the gag open reading frame. Grey areas highlight clustering of the analyzed sequences either with clade C-derived (A, C, E, and G) or with clade B-derived (B, D, and F) reference strains. Values at the nodes indicate the percent bootstraps in which the cluster to the right was supported. Bootstraps of 70% and higher only are shown.