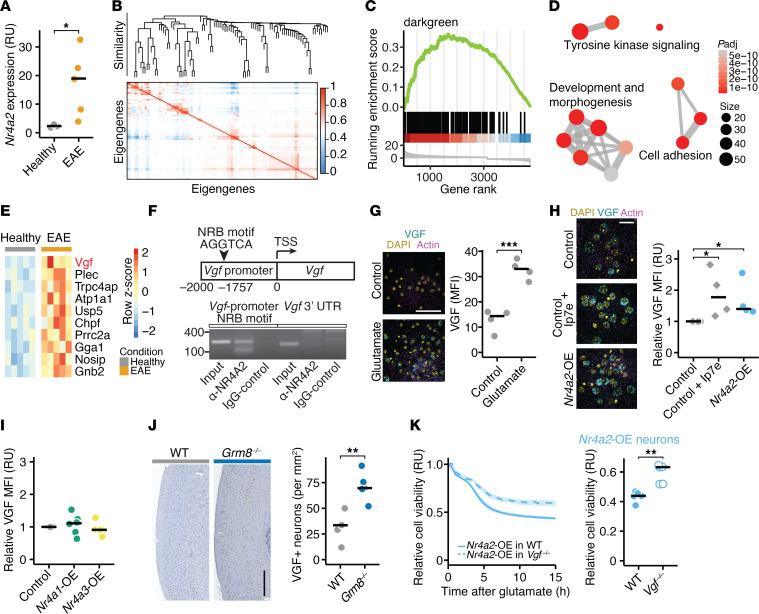
Figure 2. NR4A2-induced VGF mediates neuronal susceptibility to excitotoxicity.
(A) Nr4a2 mRNA expression in NeuN+ nuclei sorted from spinal cords of healthy mice (n = 3) and acute EAE mice (n = 5) in relative units (RU). (B) Eigengene correlation matrix and dendrogram of weighted gene correlated network analysis (WGCNA) with 502 neuron-specific transcriptome data sets. (C) Enrichment analysis of the inflamed neuronal signature (ranked gene list retrieved from ref. 9) in module “darkgreen”. (D) GO term biological process enrichment analysis of the module “darkgreen”. Size shows number of genes of GO terms, color shows significance. (E) Heatmap of top 10 genes from the module “darkgreen,” which are differently expressed in neurons during EAE. (F) Chromatin immunoprecipitation (ChIP) was performed using an antibody against NR4A2 and an IgG control from 3 pooled mouse cortices. PCR primers were designed to amplify approximately 100 nucleotides flanking the canonical nuclear receptor binding (NRB) motif in the mouse Vgf promoter (left) or the 3′ untranslated region of Vgf as control (right). (G) VGF MFI in neuronal cultures after vehicle or glutamate stimulation (n = 4 per group). Scale bar: 50 μm. (H and I) Relative VGF MFI in neurons that overexpress mScarlet (controls), Nr4a2 or were exposed to 50 nM Ip7e (H; n = 4), and neurons that overexpress Nr4a1 or Nr4a3 (I; n = 6). Data is normalized to mScarlet-overexpressing control neurons. Scale bar: 20 μm. (J) VGF+ neurons in cortices of WT and Grm8–/– mice (n = 5). Scale bar: 300 μm. (K) Cell viability (RU) of WT and Vgf–/– neurons that overexpress Nr4a2 and were exposed to glutamate (n = 5). Points represent individual experiments, additionally mean is shown. If not stated otherwise, unpaired t test with FDR correction for multiple comparisons was used. *P < 0.05, **P < 0.01, ***P < 0.001.