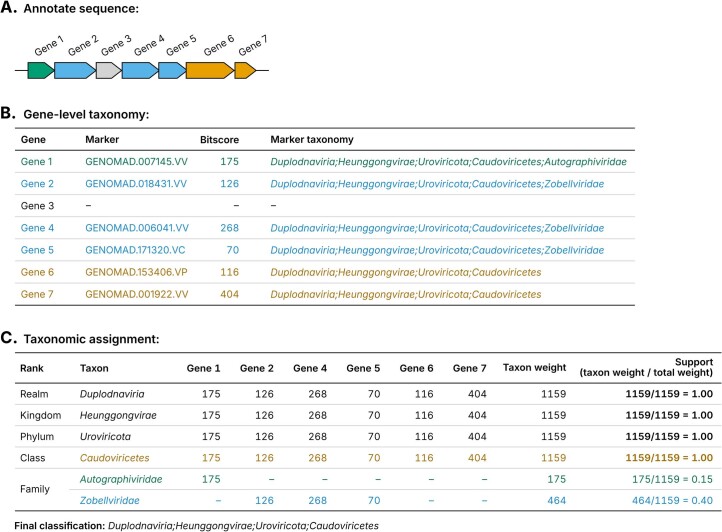
Extended Data Fig. 3. Assigning viral taxa using geNomad’s markers.
(a) To assign viral sequences to specific taxa, geNomad utilizes a best-hit approach to initially assign the genes encoded by these sequences to markers. (b) Each gene is subsequently classified based on the taxonomic lineage of the assigned marker. Different genes within the sequence might be assigned to different lineages. (c) To establish a single sequence-level taxonomy, geNomad aggregates the lineages of all the markers using a weighted majority vote approach. This approach determines the support for each taxon at each taxonomic rank by summing the bitscores of all genes assigned to that taxon. The sequence is then assigned to the most specific taxon that is supported by at least 50% of the total bitscore of the sequence.