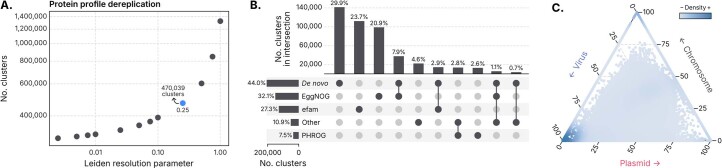
Extended Data Fig. 4. geNomad’s marker dataset was built by gathering dereplicated protein profiles from several sources and measuring their specificity to chromosomes, plasmids, and viruses.
(a) Number of protein profile clusters obtained by varying the clustering granularity (Leiden’s resolution parameter). The value chosen for dereplication (0.25) is indicated in blue. (b) UpSet plot showing the overlap of different protein profile datasets in the dereplication process. The overlap between a given pair of datasets was measured as the number of protein profile clusters that contained profiles from both. (c) Ternary plot showing the specificity of protein profiles (circles) prior to dereplication (n = 470,039). Colors represent the marker density in a region of the plot.