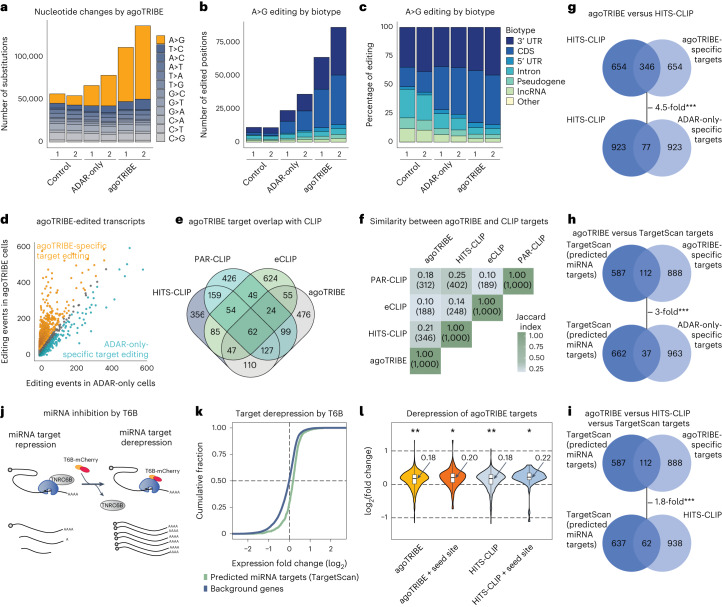
Fig. 2. agoTRIBE detects miRNA targets through RNA editing.
a, A>G editing (colored orange), indicative of ADAR2 editing, specifically increased in agoTRIBE-transfected cells compared with both control cells transfected with eGPF-Ago2 and cells transfected with the ADAR2 editing domain without Argonaute2 (ADAR-only controls). b, Editing increased specifically in 3′ UTR and coding sequences (CDS) and remained constant in other transcript types. c, As in b, but represented as percentages of editing. d, Editing in agoTRIBE-transfected versus ADAR-only-transfected cells. Each dot corresponds to one gene. Genes colored orange have substantially more editing in agoTRIBE cells while those colored light blue have substantially more editing in ADAR-only cells (Methods). Genes colored gray have comparable editing in both conditions. e, Venn diagram of top 1,000 miRNA targets reported individually by agoTRIBE, HITS-CLIP, PAR-CLIP and eCLIP. f, Jaccard similarity index values of top target sets reported by each of the four methods. g–i, Venn diagrams of overlaps between agoTRIBE and HITS-CLIP targets (g); agoTRIBE and TargetScan targets (h); and agoTRIBE, HITS-CLIP and TargetScan targets (i). j, Schematic representation of global miRNA inhibition by T6B. k, Derepression of miRNA targets predicted by TargetScan following T6B-mCherry transfection. In total, 609 TargetScan targets and 14,515 background transcripts were profiled. l, Increase in expression of agoTRIBE and HITS-CLIP targets following T6B transfection. ‘Seed site’ indicates that the transcript harbors a conserved binding site for one of the ten most abundant miRNAs in HEK-293T cells. Boxes indicate median and 25th and 75th quantiles, and whiskers indicate lowest and largest values. Significance was calculated using the Wasserstein distance (Methods); P values, left to right: 0.00025, 0.021, 0.00025, 0.012. *P < 0.05; **P < 0.001. In total, 1,000 agoTRIBE targets (112 with seed sites) and 996 HITS-CLIP targets (62 with seed sites) were profiled.