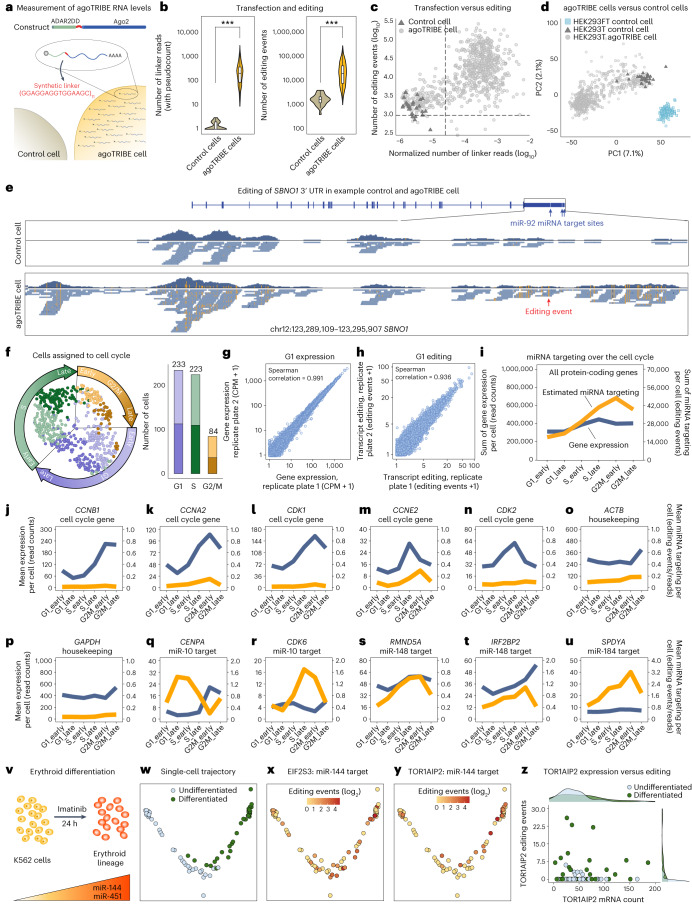
Fig. 3. miRNA targeting in single cells.
a, The agoTRIBE transcript includes an artificial linker that is detected in RNA-seq and can be used to estimate agoTRIBE levels. b, Levels of linker sequencing reads (left) and editing events (right) detected in 703 agoTRIBE-transfected cells and 26 control cells profiled by Smart-seq3 single-cell RNA-seq. Boxes indicate median and 25th and 75th quantiles, and whiskers indicate lowest and highest values. Significance was estimated using the Wilcoxon rank-sum test, with P < 0.0001. ***P < 0.0001. c, Normalized number of linker reads versus number of editing events for individual cells. The number of linker reads was normalized to the sequencing depth of each cell. Dashed lines indicate thresholds for agoTRIBE cells used for downstream analyses. d, Principal component (PC) analysis of agoTRIBE-transfected cells and control cells versus control cells from a different human embryonic kidney cell line (HEK-293FT). Cells were positioned based on their Smart-seq3 transcription profiles. e, Editing patterns of the SBNO1 transcript in a control cell versus an agoTRIBE-transfected cell. f, Overview of 540 agoTRIBE-transfected single cells assigned to cell cycle stages using their Smart-seq3 profiles. Each cell cycle stage is divided into an early and late phase (Methods). Dimensionality reduction was performed with the UMAP algorithm. g, Gene expression for cells assigned to the G1 cell cycle stage. The cells originate from two distinct replicate plates of single cells. Each dot represents one gene. h, Transcript editing for cells assigned to the G1 cell cycle stage. i, Overview of estimated overall miRNA targeting during the cell cycle. Expression values and editing events were normalized to spike-ins and to the number of cells in each cell cycle stage (Methods). j–u, Examples of transcript expression and estimated miRNA targeting during the cell cycle: CCNB1 (j), CCNA2 (k), CDK1 (l), CCNE2 (m), CDK2 (n), ACTB (o), GAPDH (p), CENPA (q), CDK6 (r), RMND5A (s), IRF2BP2 (t), SPDYA (u). Transcript expression is colored blue and miRNA targeting yellow. v, miR-144 and miR-451 increased in expression during K562 differentiation into erythroid precursor cells. w, Pseudotime trajectory of the differentiation process. Each dot indicates a single cell profiled by Smart-seq3. x,y, Editing in agoTRIBE-transfected cells during differentiation: EIF2S3 (x), TOR1AIP2 (y). z, Expression (read counts) and targeting (editing events) in TOR1AIP2. Each dot represents a single cell.