Abstract
Detecting life has driven research and exploration for centuries, but recent attempts to compile and generate a framework that summarizes life features, aimed to develop strategies for life detection missions beyond planet Earth, have disregarded a key life feature: behavior. Yet, some behaviors such as biomineralization or motility have occasionally been proposed as biosignatures to detect life. Here, we capitalize on a specific taxis’ motility behavior, magnetotaxis, to experimentally provide insights in support of behavior as an unambiguous, sensitive biosignature, and magnetic forces as a prescreening option. Using a magnetotactic bacterial species, Magnetospirillum magneticum, we conducted a lab sensitivity experiment comparing PCR with the hanging drop behavioral assay, using a dilution series. The hanging drop behavioral assay visually shows the motility of MTB toward magnetic poles. Our findings reveal that the behavioral assay exhibits higher sensitivity in the detection of M. magneticum when compared to the established PCR protocol. While both methods present similar detection sensitivities at high concentrations, at ≥ 10–7 fold dilutions, the behavioral method proved more sensitive. The behavioral method can detect bacteria even when samples are diluted at 10–9. Comparable results were obtained with environmental samples from the Hula Valley. We propose behavioral cues as valuable biosignatures in the ongoing efforts of life detection in unexplored aquatic habitats on Earth and to stimulate and support discussions about how to detect extant life beyond Earth. Generic and robust behavioral assays can represent a methodological revolution.
Keywords: Life detection, Behavior, Magnetotaxis, Magnetospirillum magneticum, Hanging drop assay, PCR
Subject terms: Evolution, Microbiology
Introduction
Detecting life at remote locations, under extreme conditions, past and present, on Earth and beyond, has driven research and exploration for centuries1. In the past fifty years, our understanding of biological processes and the limits of life on Earth have greatly improved thanks to technological advances (in molecular biology and remote sensing), the discovery of extreme ecosystems like chemolithotrophic communities on the ocean floor2, and the exploration of the solar system by interplanetary missions3. Life-detection efforts are once again a high research priority if they ever stopped4.
Recent attempts to compile and generate a Ladder that summarizes life features, aimed to stimulate and generate discussion regarding life detection beyond planet Earth5, have disregarded a key life feature: behavior. Behavior is the internally coordinated responses of whole organisms to internal and external stimuli6. Fundamentally, behavior is a biosignature of evolutionary processes, a coordinated living organism adaptation. The exclusion of behavior as a key feature of life is an unfortunate omission, since behavioral responses in large organisms would readily be viewed as evidence for life. What’s missing is recognition that behavior can be used to detect microbial life. Recognizing behavior has typically been challenging in microbes, even though microbially induced biogenic structures, microbiolites7, and individual cell motility8 have occasionally been considered as potential biosignatures of extraterrestrial microbial life. These difficulties have led to behavior being overlooked in the recently proposed Ladder to search for extraterrestrial life.
Traditionally, when trying to detect microorganisms, multiple criteria pertaining to the technique (i.e., sensitivity or repeatability) or measuring context (i.e., limits of detection, specificity, or compatibility) come into play. Behavioral methods were previously widely used in microbiology (e.g., motility or chemotactism, just to name a few9–11). In recent years, the polymerase chain reaction (PCR) has become the cornerstone of molecular biology to detect and identify microbial life. PCR replicates segments of DNA/RNA in a semiconservative way with generally good detection limits. Multiple studies in the biomedical diagnostic field have studied the sensitivity of PCR in bacterial pathogens, as well as real-time PCR and multiplex PCR sensitivity, with studies reporting a detection limit of 10 genome copies per PCR reaction on extracted genomic DNA12. Nevertheless, studies comparing culturing and molecular assay methods for quantitative detection of bacteria give discordant results and higher sensitivity of culturing techniques (i.e., fecal stool samples13 or environmental samples14,15).
Behavioral methods can serve as a good complementary method to molecular techniques. To target amplification, one needs to have some previous knowledge regarding what they are looking for (specificity of the primer(s)). In behavioral methods, the selection procedure is the method. It exploits a pre-existing adaptation that will differentiate the organisms from their abiotic environment. In most cases, behavioral methods make it hard to predict what and if will be detected. With a bit of thought, one can specify a behavioral assay to a specific adaptation. Similarly, some behavior-based methods are specific for certain groups of organisms, but again these specificities are based on environmental adaptation rather than a phylogenetic indicator. Such is the case for the magnetic response of magnetotactic bacteria (from here on MTB).
MTB is a polyphyletic group of prokaryotes that produce magnetosomes, organelles that allow detection and orientation of the MTB body along the Earth’s geomagnetic field lines16. They are ubiquitous in freshwater bodies, saline and alkaline lakes, hot springs, and hemipelagic and deep-sea sediment environments4,17–23. In addition, MTB have also been the focus of astrobiology, in the search for extraterrestrial life4. While the adaptive value of bacterial magnetotaxis is still under debate24,25, the leading hypothesis is that magnetosomes serve as a navigational device allowing MTB to orient and migrate along geomagnetic field lines, thus reducing the cost of movement from a three-dimensional space to a one-dimensional one26,27. Generally, MTB are grouped by their magneto-aerotaxis behaviors in two main groups: polar, in which the cells swim persistently parallel or antiparallel to the magnetic field, and axial, in which cells swim in either direction along the magnetic field lines with frequent, spontaneous reversals25,28. However, several magneto-aerotaxis behaviors28, as well as other mechanisms not exclusively dependent on oxygen gradients such as phototaxis29,30 and chemotaxis31, have been described in different MTB strains.
The distinctive behavior of this group of gram-negative bacteria (magnetic sensitivity) allows for a behavioral assay (e.g., the “hanging drop” method) unconstrained by phylogeny/taxonomic categorization or growth dynamics32. The hanging drop assay is a well-established method for microbial motility testing of living organisms. This long-used behavioral method that does not require knowledge of the species-specific taxonomy, a specific DNA sequence, or any specific metabolic signature—just a simple “old school” behavioral assay.
Here, we carry out a prospective comparison of PCR assays and the hanging drop technique (HDT) in the detection of Magnetospirillum magneticum, an MTB species that serves as a model organism to study MTB and magnetism in general. The study aims to address the sensitivity of both PCR and HDT in identifying magnetotactic bacteria at low concentrations. These simple experiments demonstrate, once again, the power of behavioral techniques to detect microbial life. We propose to revisit the life detection Ladder5, incorporating behavioral methods.
Methods
Strain and culture conditions
M. magneticum strain AMB-1 was purchased from ATCC (ATCC 700264). M. magneticum exhibits axial magneto-aerotaxis, Cultures were grown in ATCC Medium 1653 (Revised Magnetic spirillum growth Medium (MSGM)) prepared according to the ATCC protocol without Resazurin. Bacteria were maintained in batch culture using 15 ml conical centrifuge tubes at 27 °C. Populations were transferred to fresh MSGM medium every four days. Before conducting the experiments, M. magneticum cultures were evaluated for non-contamination using the hanging drop technique (see below).
Sensitivity assays
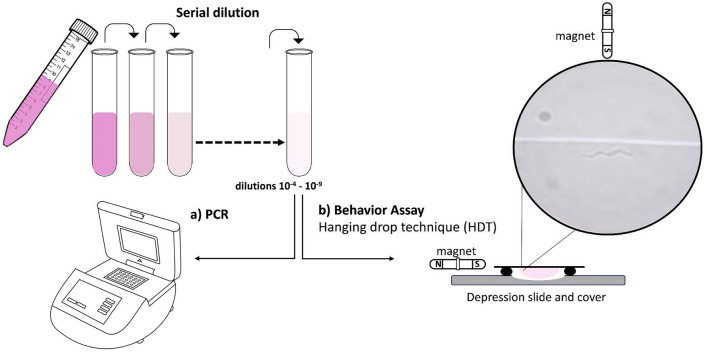
A dilution row with dilution groups between 10–4 until 10–10 was the base of this experiment (Fig. 1). 1 μl of a grown population of M. magneticum was diluted in 9.999 ml of MSGM, providing a dilution stock of 10–4. A step-wise ten-fold series of dilutions of the M. magneticum population was used up to 10–10 dilutions. Paired samples of 1 ml were drawn from each final dilution. One sample was used for visual examination using the HDT and the second was frozen at − 20 °C for later DNA extraction and PCR amplification. The time of freezing the second sample was correlated with the moment of drawing 10 μl from the first tube dedicated to the HDT. Characterization of each dilution group was repeated four times. HDT enables us to test motility of living M. magneticum cells. After exposure to the magnet’s magnetic field at one edge of the drop, we can count the cells that swam to and accumulated at the edge closest to the magnet, the opposite edge, and around the drop perimeter.
Figure 1.
Experimental design schematic diagram. Serial dilutions of M. magneticum cultures grown in revised MSGM were assessed. Aliquots from dilutions between 10–4 until 10–10 were used for DNA extraction and PCR amplification (a). In parallel, aliquots were visually examined with HDT and optical microscopy (b). M. magneticum cells responded via taxis towards a magnet’s magnetic field. The microphotograph shows a cell of M. magneticum swimming towards the magnet's south pole at a dilution 10–8 (×100 objective).
For conducting the HDT, 1 ml dilution samples in 1.5 ml tubes were placed above the magnetic north of a stirring magnet, held upright by a rack with the tip of the 1.5 ml tube pointing downwards to the magnet. For each dilution row and population, two 1 ml samples were visually examined with the hanging drop method. The first sample was exposed to the magnet for 40 min, and the second for 50 min. After the set exposure time, 10 µl of the sample’s liquid was drawn from the tube’s tip, and a drop was formed on a hydrophobic cover slip. Coverslips were flipped upside down and placed on a glass slide with a ring, allowing the drop to hang loosely without touching the surface underneath. The sample was examined using a micros MCX51, upright microscope with an Olympus sc100 camera and further exposure to a stirring magnet pointing towards the hanging drop from the side. Live M. magneticum orient and move in either direction along the magnetic field lines accumulating at the drop border of the drop (Suppl. Video 1). M. magneticum presence at the border of the drop next to the magnet, and in the drop perimeter, was visually assessed. In low-concentration samples (10–7 and lower), single bacteria were counted, with emphasis on both the magnetic north and south sides of the drop. Dead M. magneticum cells tend to burst open; even though their shapes were still visible, they weren’t counted to exclude confusion with potential floating particles in the medium. Immobile but intact M. magneticum cells were counted. At high concentrations, instead of counting single cells, the overall proportion of area covered between each side of the drop (north vs south) was visually assessed, see the percentage in Table 1, concentrations 10–4–10–6.
Table 1.
Primer details.
| Primer pair | Sequence (5′ → 3′) |
|---|---|
| MamQ |
Fw: CTGCAACGGGTGAAGCAGT Rev: TCCTGCGCATGGTTGAGAG |
| MamT |
Fw: TCGGACTGGGACTCTATTGGGA Rev: CTTTTCCACAGGCACCTTGACC |
DNA extraction and PCR amplification
DNA extraction was conducted using an extraction kit (Invitrogen Purelink Microbiome DNA Purification Kit, Thermo Fisher), following the manufacturer's protocol. Following extraction, the processed samples were then subjected to PCR amplification and finally stored at – 20 °C for future use.
MamQ and MamT primers (Table 1) were used for the amplification of 2 mam genes (MamQ and MamT) reported to be involved in the formation of the magnetosome. Primer sequences were designed following Lefèvre et al.33, with slight modifications (Table 1).
PCR amplification was performed using a total reaction volume of 25 µl, consisting of 2 µl of extracted DNA and 23 µl of PCR mix: 12 µl PrimeSTAR Max PCR mix (Takara Bio, Europe) or Phanta Flash Master Mix (Vazyme, Nanjing, PRC), 2 µl of forward and 2 µl of reverse primers and 7 µl DDW. The program protocol included an initial denaturation stage at 94 °C for 2 min, followed by 35 cycles of 30 s of denaturation at 94 °C, annealing for 30 s at 55.5 °C, and an elongation stage for 15 s at 72 °C. Afterward a final extension stage of 7 min at 72 °C was used. At the end of the reaction, the samples were held at 4 °C. A high concentration of MTB served as positive control and DDW served as a negative control in the PCR reaction. The results of a PCR reaction were visualized using gel electrophoresis (1.5% Agarose in TBE buffer). Electrophoresis gels were loaded with 5–10 μl of purified PCR product mixed with 1–2 μl tracking dye. A DNA ladder was used as a molecular-weight size marker.
Statistical Analyses
Statistical analysis was done using JMP16 (SAS Inc). To examine the binomial probability of between-group detection differences in detecting n positive out of N samples, we used a generalized linear model with binomial distribution and a logit link function. The predictor variable was HDT vs. PCR and the dependent variable was n positive out of N samples in each dilution. In addition, we examined the binomial difference in detection in all low-concentration dilutions merged (10–7–10–9).
Results
HDT was significantly more sensitive regarding the detection of lower concentrations of M. magneticum in single-strain culture compared with the PCR detection using both MamQ and MamT primers (X2 = 19.74, p < 0.0001). Up to dilutions of 10–6 fold, using the HDT, M. magneticum densities were ≥ 50 cells/10 µl both swimming with and opposite to the induced magnetic field (Table 2). Between 10–7 and 10–8 dilution factors, cellular densities lower than 50 cells and up to 3 cells were positively detected with the HDT, with a mix of positive and negative detection results from PCR. When we obtained PCR-positive results, the results were positive with both MamQ and MamT primers. Dilutions of 10–9 fold were only positive using the HDT suggesting a limit of detection for the PCR technique.
Table 2.
M. magneticum densities, behavior with respect to the magnetic field, and detection results using HDT and PCR.
| Dilution | HDT | PCR | Significance by group* | ||||
|---|---|---|---|---|---|---|---|
| North seeking bacteria | South seeking bacteria | Total count of bacteria | [pos./neg.] [1,0] | MamQ [pos./neg.] [1,0] |
MamT [pos./neg.] [1,0] |
||
| 10–4 | N/A | N/A | ≥ 50 | 1 | 1 | 1 | |
| 10–4 | N/A | N/A | ≥ 50 | 1 | 1 | 1 | |
| 10–4 | 90% | 10% | ≥ 50 | 1 | 1 | 1 | |
| 10–4 | 90% | 10% | ≥ 50 | 1 | 1 | 1 | |
| 4/4 | 4/4 | 4/4 | P > 0.5 | ||||
| 10–5 | 40% | 60% | ≥ 50 | 1 | 1 | 1 | |
| 10–5 | 50% | 50% | ≥ 50 | 1 | 1 | 1 | |
| 10–5 | 90% | 10% | ≥ 50 | 1 | 1 | 1 | |
| 10–5 | 90% | 10% | ≥ 50 | 1 | 1 | 1 | |
| 4/4 | 4/4 | 4/4 | P > 0.5 | ||||
| 10–6 | 50% | 50% | ≥ 50 | 1 | 1 | 1 | |
| 10–6 | 50% | 50% | ≥ 50 | 1 | 1 | 1 | |
| 10–6 | 80% | 20% | ≥ 50 | 1 | 1 | 1 | |
| 10–6 | 70% | 30% | ≥ 50 | 1 | 1 | 1 | |
| 4/4 | 4/4 | 4/4 | P > 0.5 | ||||
| 10–7 | N/A | N/A | 4 | 1 | 0 | 0 | |
| 10–7 | N/A | N/A | 8 | 1 | 0 | 0 | |
| 10–7 | N/A | N/A | 35 | 1 | 1 | 1 | |
| 10–7 | N/A | N/A | 20 | 1 | 1 | 1 | |
| 10–7 | 17 | 22 | 39 | 1 | 0 | 0 | |
| 10–7 | 40 | 4 | 44 | 1 | 0 | 0 | |
| 6/6 | 2/6 | 2/6 | 0.0057 | ||||
| 10–8 | 3 | 3 | 6 | 1 | 0 | 0 | |
| 10–8 | 8 | 1 | 9 | 1 | 0 | 0 | |
| 10–8 | 3 | 0 | 3 | 1 | 1 | 1 | |
| 10–8 | 3 | 1 | 4 | 1 | 1 | 1 | |
| 4/4 | 2/4 | 2/4 | 0.0632 | ||||
| 10–9 | 1 | 0 | 7 | 1 | 0 | 0 | |
| 10–9 | 3 | 0 | 3 | 1 | 0 | 0 | |
| 10–9 | 3 | 1 | 4 | 1 | 0 | 0 | |
| 10–9 | 1 | 0 | 1 | 1 | 0 | 0 | |
| 4/4 | 0/4 | 0/4 | 0.0009 | ||||
| 10–10 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 10–10 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 10–10 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 10–10 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 0/4 | 0/4 | 0/4 | P > 0.5 | ||||
At 10–9 fold dilutions, HDT can detect up to 1 single observable cell, whereas the PCR method is not able to detect the presence of the MTB. P values indicate the difference using a binomial distribution test with a logit link function. When examining the sensitivity difference in all these lower concentrations together, these differences are highly significant (X2 = 19.74, p < 0.0001). N/A refers to not being assessed.
Discussion
One of the most prominent phenotypes resulting from adaptation is an organism’s behavior34. Here, we demonstrated that a simple, non-species-specific, behavioral assay can be significantly more sensitive in detecting microbes (in this case, MTB) than the standard PCR assay. We raise awareness about using behavior as a missing biosignature of “Darwinian evolution” in the Ladder of Life Detection paradigm.
Comprehensive studies utilizing multiple detection methods are required to identify terrestrial and extraterrestrial microorganisms (35,36). In this study, we examined two detection methods, a PCR method and the HDT, using commercial M. magneticum under lab conditions and with species-specific well-designed primers. Theoretically, PCR detection methods with specific primers presented a higher detection power for M. magneticum than HDT. DNA is amplified easily during PCR despite the cell's state. The process of dilution and the HDT exposes the M. magneticum to oxygen, which kills a portion of the MTB. While dead or alive status should not affect PCR, dead cells will not actively swim toward the magnetic field and will not be detected with the HDT. Moreover, M. magneticum is a laboratory-adapted strain under “relaxed selection” for magnetism37. The HDT relies on the cells actively swimming towards or against the imposed magnetic field, whereas a lack of behavioral response will not affect the PCR results. However, despite the above-mentioned, the behavioral method presented higher sensitivity when compared with our PCR method. Our results indicate a higher depth and accuracy of the HDT in identifying low concentrations of M. magneticum. This study aims to highlight the sensitivity of the behavioral technique and its potential for life detection, using gene amplification by PCR as a reference. Other molecular techniques, such as whole genome sequencing, shotgun metagenomics, or single-cell sequencing, will provide different results or detection outcomes at low dilutions. While the advantages and limitations of each method vary, every amplification method has its weaknesses38–40. Here we used the standard PCR under very favorable conditions as a simple and robust test, but we cannot rule out that more sensitive methods theoretically could also detect bacteria in low concentration. However, our results, as they are, demonstrate the overlooked power and sensitivity of behavioral assays.
Furthermore, in a preliminary proof of concept (see supplemental materials), we took the two methods out of the lab and to the field. The sensitivity of the HDT substantially outweighs our PCR method. We examined forty-one environmental samples, obtained from shallow wetlands in the Hula Valley, north Israel, which were all positive for MTB with the HDT. Visual identification of the magnetic response of different species and morphological examination revealed mainly morphologically Magnetotactic cocci, but at least six morphologically distinct morphotypes including Magnetotactic spirillum, were identified in various samples. None were amplified with the various sets of primers that we examined (see supplemental materials). Obviously, the PCR’s of the environmental samples could be improved following sequencing and the creation of more specific primers and improved protocols. However, this demonstrates the generality of the behavioral method, and concretely, the sensitivity of HDT, makes it a more reliable screening technique to identify specific species from field samples. Problems of PCR sensitivity, specificity, and robustness have previously been flagged in environmental samples such as soil, water, leaves, or insects40–43, and our findings support these concerns. Given the ubiquity of genomic approaches, it is imperative to understand the applications, limitations, and basic principles of genetic approaches to identify microbial life. DNA sequences provide documentary evidence of biological presence and evolutionary past44, but their capabilities to detect microbial life are still far from being realized.
Behavioral methods exploit adaptive behaviors allowing generalization and wider applicability with the trade-offs of losing specificity and accuracy. These techniques don’t require specific knowledge of a DNA sequence, species-specific structure, or system’s function. Specifically, MTB as a group displays high sensitivity to the magnetic field, exhibiting bidirectional locomotion in the case of axial MTB. Magnetotaxis’ evolutionary advantages are not well understood. This behavior has been explained in terms of energy efficiency mainly in relation to vertical orientation towards optimal conditions of oxygen and reduction potential26. Using magnetic force, we could attract and observe multiple species, if present, enabling the discovery of MTB. Recent examples are the discovery of MTB in hydrothermal vents in the deep sea45, marine sediments (36,46,47), or the report of mutualistic symbiosis between an MTB and a marine protist48. Therefore, among the multiple examples of microbial behavioral biology, we could state that magnetotaxis is highly specific in the detection of a particular group of microorganisms that express this function.
Taking this discussion to astrobiology, magnetotaxis and MTB are regarded as especially important in the search for extraterrestrial life49. Research has been conducted to test the effect of microgravity on MTB50. Lately, there has been a boom in space exploration and specifically a growing interest in extraterrestrial MTB4. The recent discovery of an MTB from a hot hydrothermal vent in the deep sea45 has pointed out MTB as promising biosignatures to detect extraterrestrial life where hydrothermal activities ever occurred, i.e., Mars or icy moons. Moreover, notwithstanding the controversy from the previously reported magnetotactic bacteria fossils in the Martian meteorite Allan Hills 84001, several biosignatures have been described and suggested as promising landmarks to look for beyond Earth1,4.
When searching for life, the value of any insight is greatest when supported in combination with other approaches. However, in the quest for the detection of life, it seems that behavioral ecology has been somewhat left out. Instead of behavioral ecology, measurements of thermodynamics, shapes, and waste are well-researched1,4. This is largely justified as many times, extraterrestrial life exploration examines remnants or signatures of past life forms (e.g. Mars). As previous comparative studies for life detection concluded, no single method presents advantages in all respects, nor can a single method realistically unilaterally conclude the discovery of extraterrestrial, or even terrestrial, life35. Our simple experiment demonstrates the appeal of using a behavioral method when examining extant life. We propose that in addition to the standard methods, we can ask ourselves what would be adaptive in these environments, and what behavioral assays may be developed accordingly (e.g. chemotaxis, magnetotaxis, phototaxis); as we demonstrated, their sensitivity might be surprising.
The generic nature of behavioral techniques, along with their robustness, makes behavioral biology the ultimate proof of life51. We propose behavioral assays as the missing biosignature of evolution in the Ladder of Life Detection1. Using the needle in a haystack metaphor example: if you use a magnet the MTB will come to you.
Supplementary Information
Author contributions
M.T., Y.V. and B.B. participated in the conceptualization, D.Z. and M.L. performed the experiments, Y.V. Funding acquisition, E.N., S.T. and O.K contributed with Methodology and Resources, Y.V., M.T. and B.B. Supervision, B.B. (lead) and Y.V. (supported) Writing—original draft and D.Z., M.L., E.N., S.T., O.K., M.T., Y.V. and B.B. contributed to Writing—review and editing.
Funding
This research was supported by the Israel Science Foundation (Grant No. 1350/21) to Y.V., the Hula Research Center is supported by KKL-JNF and Tel-Hai College in a joint venture. B.B. is supported by a PPFP fellowship from the University of Minnesota.
Data availability
Data and materials availability: All data are available in the manuscript or the supplementary materials. Raw images and videos presented and discussed in this paper can be found in Zenodo, 10.5281/zenodo.11356036.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Michael Travisano, Yoni Vortman and Beatriz Baselga-Cervera.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-69942-y.
References
- 1.Neveu, M., Hays, L. E., Voytek, M. A., New, M. H. & Schulte, M. D. The ladder of life detection. Astrobiology18, 1375–1402 (2018). 10.1089/ast.2017.1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corliss, J. B. et al. Submarine thermal springs on the Galápagos rift. Science203, 1073–1083 (1979). 10.1126/science.203.4385.1073 [DOI] [PubMed] [Google Scholar]
- 3.Dick, S. J. NASA and the search for life in the universe. Endeavour30, 71–75 (2006). 10.1016/j.endeavour.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 4.Shen, J. et al. Renaissance for magnetotactic bacteria in astrobiology. ISME J.17, 1526–1534 (2023). 10.1038/s41396-023-01495-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.NASA Astrobiology. https://astrobiology.nasa.gov/research/life-detection/ladder/.
- 6.Levitis, D. A., Lidicker, W. Z. & Freund, G. Behavioural biologists do not agree on what constitutes behaviour. Anim. Behav.78, 103–110 (2009). 10.1016/j.anbehav.2009.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, M. A. et al. Deciphering biosignatures in planetary contexts. Astrobiology19, 1075–1102 (2019). 10.1089/ast.2018.1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadeau, J., Lindensmith, C., Deming, J. W., Fernandez, V. I. & Stocker, R. Microbial morphology and motility as biosignatures for outer planet missions. Astrobiology16, 755–774 (2016). 10.1089/ast.2015.1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adler, J. Chemoreceptors in bacteria: Studies of chemotaxis reveal systems that detect attractants independently of their metabolism. Science166, 1588–1597 (1969). 10.1126/science.166.3913.1588 [DOI] [PubMed] [Google Scholar]
- 10.Berg, H. C. Bacterial behaviour. Nature254, 389–392 (1975). 10.1038/254389a0 [DOI] [PubMed] [Google Scholar]
- 11.Madigan, M. T., Clark, D. P., Stahl, D. & Martinko, J. M. Brock Biology of Microorganisms 13th Edition. In Basic Principles of Microbiology (ed. 13th, 2012; https://www.academia.edu/42749825/Brock_Biology_of_Microorganisms_13th_Edition), pp. 1–84.
- 12.Sabat, G., Rose, P., Hickey, W. J. & Harkin, J. M. Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl. Environ. Microbiol.66, 844–849 (2000). 10.1128/AEM.66.2.844-849.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hapuarachchi, C. T., Jeffery, K. J. M. & Bowler, I. C. J. W. Stool PCR may not be a substitute for enrichment culture for the detection of salmonella. J. Med. Microbiol.68, 395–397 (2019). 10.1099/jmm.0.000923 [DOI] [PubMed] [Google Scholar]
- 14.Corless, C. E. et al. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol.38, 1747–1752 (2000). 10.1128/JCM.38.5.1747-1752.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morán, F. et al. The challenge of environmental samples for PCR detection of phytopathogenic bacteria: A case study of citrus huanglongbing disease. Agronomy11, 10 (2020). 10.3390/agronomy11010010 [DOI] [Google Scholar]
- 16.Shively, J. M., Cannon, G. C., Heinhorst, S., Bryant, D. A., DasSarma, S., Bazylinski, D., Preiss, J., Steinbüchel, A., Docampo, R. & Dahl, C. Bacterial and archaeal inclusions. In Encyclopedia of Life Sciences (Wiley, ed. 1, 2011; 10.1002/9780470015902.a0000302.pub3).
- 17.Bazylinski, D. & Lefèvre, C. Magnetotactic bacteria from extreme environments. Life3, 295–307 (2013). 10.3390/life3020295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bazylinski, D. A. & Frankel, R. B. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol.2, 217–230 (2004). 10.1038/nrmicro842 [DOI] [PubMed] [Google Scholar]
- 19.Busigny, V. et al. Mass collection of magnetotactic bacteria from the permanently stratified ferruginous Lake Pavin, France. Environ. Microbiol.24, 721–736 (2022). 10.1111/1462-2920.15458 [DOI] [PubMed] [Google Scholar]
- 20.Dong, Y. et al. The detection of magnetotactic bacteria in deep sea sediments from the east Pacific Manganese Nodule Province. Environ. Microbiol. Rep.8, 239–249 (2016). 10.1111/1758-2229.12374 [DOI] [PubMed] [Google Scholar]
- 21.Lefèvre, C. T., Frankel, R. B., Pósfai, M., Prozorov, T. & Bazylinski, D. A. Isolation of obligately alkaliphilic magnetotactic bacteria from extremely alkaline environments. Environ. Microbiol.13, 2342–2350 (2011). 10.1111/j.1462-2920.2011.02505.x [DOI] [PubMed] [Google Scholar]
- 22.Petermann, H. & Bleil, U. Detection of live magnetotactic bacteria in South Atlantic deep-sea sediments. Earth Planet. Sci. Lett.117, 223–228 (1993). 10.1016/0012-821X(93)90128-V [DOI] [Google Scholar]
- 23.Stolz, J. F., Chang, S.-B.R. & Kirschvink, J. L. Magnetotactic bacteria and single-domain magnetite in hemipelagic sediments. Nature321, 849–851 (1986). 10.1038/321849a0 [DOI] [Google Scholar]
- 24.Natan, E. & Vortman, Y. The symbiotic magnetic-sensing hypothesis: Do Magnetotactic Bacteria underlie the magnetic sensing capability of animals?. Mov. Ecol.5, 22 (2017). 10.1186/s40462-017-0113-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barber-Zucker, S., Keren-Khadmy, N. & Zarivach, R. From invagination to navigation: The story of magnetosome-associated proteins in magnetotactic bacteria. Protein Sci. Publ. Prot. Soc.25, 338–351 (2016). 10.1002/pro.2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frankel, R. B., Bazylinski, D. A., Johnson, M. S. & Taylor, B. L. Magneto-aerotaxis in marine coccoid bacteria. Biophys. J.73, 994–1000 (1997). 10.1016/S0006-3495(97)78132-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frankel, R. B., Williams, T. J. & Bazylinski, D. A. Magneto-aerotaxis. In Magnetoreception and Magnetosomes in Bacteria (ed. Schüler, D.) 1–24 (Springer, Berlin, 2007). 10.1007/7171_2006_036. [Google Scholar]
- 28.Lefèvre, C. T. et al. Diversity of magneto-aerotactic behaviors and oxygen sensing mechanisms in cultured magnetotactic bacteria. Biophys. J.107, 527–538 (2014). 10.1016/j.bpj.2014.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, C., Ma, Q., Jiang, W. & Song, T. Phototaxis in the magnetotactic bacterium Magnetospirillum magneticum strain AMB-1 is independent of magnetic fields. Appl. Microbiol. Biotechnol.90, 269–275 (2011). 10.1007/s00253-010-3017-1 [DOI] [PubMed] [Google Scholar]
- 30.Shapiro, O. H., Hatzenpichler, R., Buckley, D. H., Zinder, S. H. & Orphan, V. J. Multicellular photo-magnetotactic bacteria. Environ. Microbiol. Rep.3, 233–238 (2011). 10.1111/j.1758-2229.2010.00215.x [DOI] [PubMed] [Google Scholar]
- 31.Alexandre, G., Greer-Phillips, S. & Zhulin, I. B. Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev.28, 113–126 (2004). 10.1016/j.femsre.2003.10.003 [DOI] [PubMed] [Google Scholar]
- 32.Bazylinski, D. A., Lefèvre, C. T. & Schüler, D. Magnetotactic bacteria. In The Prokaryotes: Prokaryotic Physiology and Biochemistry (eds Rosenberg, E. et al.) 453–494 (Springer, 2013). 10.1007/978-3-642-30141-4_74. [Google Scholar]
- 33.Lefèvre, C. T. et al. Insight into the evolution of magnetotaxis in Magnetospirillum spp., based on mam gene phylogeny. Appl. Environ. Microbiol.78, 7238–7248 (2012). 10.1128/AEM.01951-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies, N. B., Krebs, J. R. & West, S. A. An Introduction to Behavioural Ecology (Wiley, 2012). [Google Scholar]
- 35.Enya, K., Yamagishi, A., Kobayashi, K. & Yoshimura, Y. Comparative study of methods for detecting extraterrestrial life in exploration mission of Mars and the solar system. Life Sci. Space Res.34, 53–67 (2022). 10.1016/j.lssr.2022.07.001 [DOI] [PubMed] [Google Scholar]
- 36.Suzuki, S. et al. Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM. Commun. Biol.5, 707 (2022). 10.1038/s42003-022-03668-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ullrich, S., Kube, M., Schübbe, S., Reinhardt, R. & Schüler, D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J. Bacteriol.187, 7176–7184 (2005). 10.1128/JB.187.21.7176-7184.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capron, A. et al. In situ processing and efficient environmental detection (iSPEED) of tree pests and pathogens using point-of-use real-time PCR. PLoS ONE15, e0226863 (2020). 10.1371/journal.pone.0226863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahsanuddin, S. et al. Assessment of REPLI-g multiple displacement whole genome amplification (WGA) techniques for metagenomic applications. J. Biomol. Tech.28(2), 96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ordóñez, C. D. & Redrejo-Rodríguez, M. DNA polymerases for whole genome amplification: Considerations and future directions. Int. J. Mol. Sci.24(11), 9331 (2023). 10.3390/ijms24119331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franco-Duarte, R. et al. Advances in chemical and biological methods to identify microorganisms—From past to present. Microorganisms7, 130 (2019). 10.3390/microorganisms7050130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saingam, P., Li, B. & Yan, T. Use of amplicon sequencing to improve sensitivity in PCR-based detection of microbial pathogen in environmental samples. J. Microbiol. Methods149, 73–79 (2018). 10.1016/j.mimet.2018.05.005 [DOI] [PubMed] [Google Scholar]
- 43.Smith, C. J. & Osborn, A. M. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology: Application of Q-PCR in microbial ecology. FEMS Microbiol. Ecol.67, 6–20 (2009). 10.1111/j.1574-6941.2008.00629.x [DOI] [PubMed] [Google Scholar]
- 44.Hughes, A. L. The origin of adaptive phenotypes. Proc. Natl. Acad. Sci.105, 13193–13194 (2008). 10.1073/pnas.0807440105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakano, S. et al. Bullet-shaped magnetosomes and metagenomic-based magnetosome gene profiles in a deep-sea hydrothermal vent chimney. Front. Microbiol.14, 1174899 (2023). 10.3389/fmicb.2023.1174899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui, K. et al. Characterization and diversity of magnetotactic bacteria from sediments of Caroline Seamount in the Western Pacific Ocean. J. Oceanol. Limnol.39, 2027–2043 (2021). 10.1007/s00343-021-0029-x [DOI] [Google Scholar]
- 47.Liu, J. et al. Bacterial community structure and novel species of magnetotactic bacteria in sediments from a seamount in the Mariana volcanic arc. Sci. Rep.7, 17964 (2017). 10.1038/s41598-017-17445-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monteil, C. L. et al. Ectosymbiotic bacteria at the origin of magnetoreception in a marine protist. Nat. Microbiol.4, 1088–1095 (2019). 10.1038/s41564-019-0432-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKay, C. P., Friedmann, E. I., Frankel, R. B. & Bazylinski, D. A. Magnetotactic bacteria on earth and on mars. Astrobiology3, 263–270 (2003). 10.1089/153110703769016361 [DOI] [PubMed] [Google Scholar]
- 50.Urban, J. E. Adverse effects of microgravity on the magnetotactic bacterium Magnetospirillum magnetotacticum. Acta Astronaut.47, 775–780 (2000). 10.1016/S0094-5765(00)00120-X [DOI] [PubMed] [Google Scholar]
- 51.Wallace, R. A., Sanders, G. P. & Ferl, R. J. Biology, the Science of Life 3rd edn. (HarperCollins, 1991). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and materials availability: All data are available in the manuscript or the supplementary materials. Raw images and videos presented and discussed in this paper can be found in Zenodo, 10.5281/zenodo.11356036.