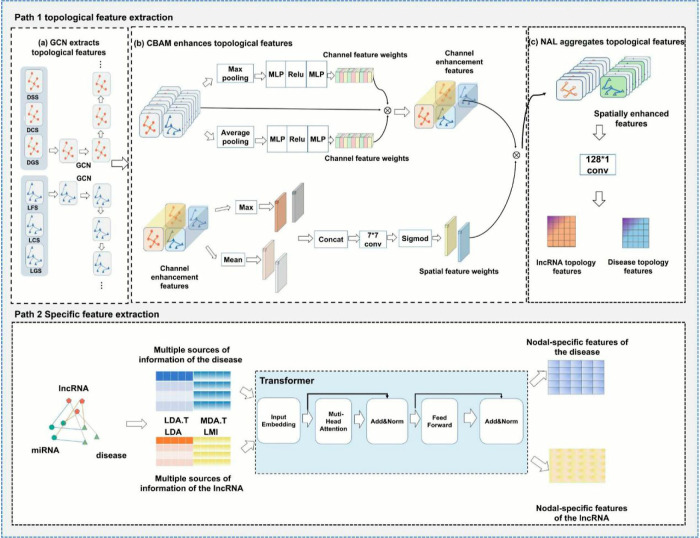
Figure 2.
Presents the framework of the model, which consists of two parts. The first part is the node topology feature extraction network, which is responsible for extracting the node topology features. The node features from various convolutional layers of the GCN are enhanced using CBAM for channelwise and spatialwise feature enhancement. Then, the aggregated features are passed through the NAL for multichannel convolutional dimension reduction. The second part is the node topology feature residual network, which utilizes the heterogeneous association information between lncRNAs and diseases, along with the Transformer, to extract node-specific features. This network focuses on capturing the specific features of the nodes related to lncRNAs and diseases.