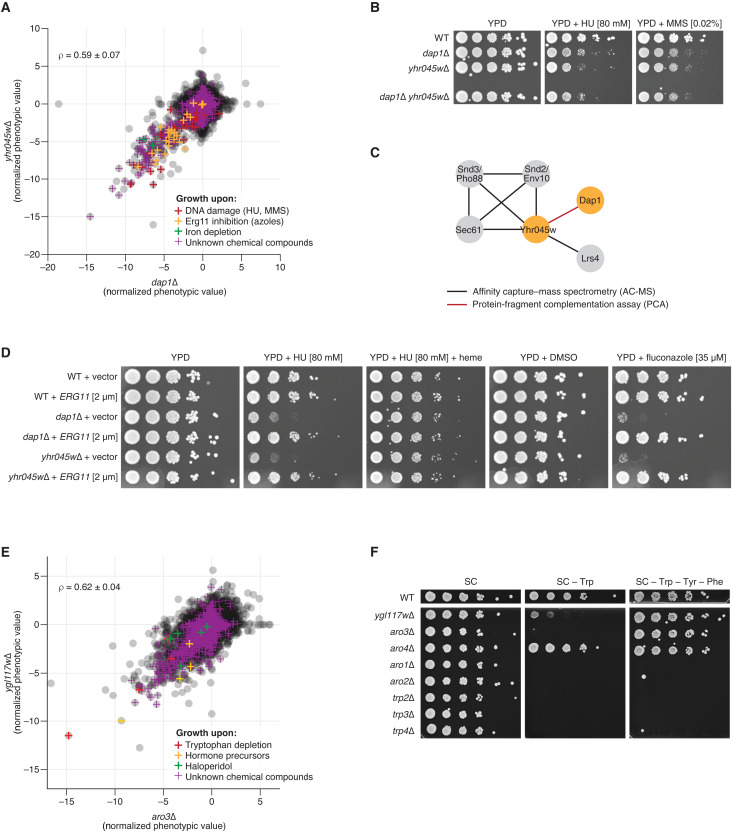
Fig. 4. Functional validation of YHR045W and YGL117W.
(A) The similarity of the phenotypic profiles of yhr045wΔ and dap1Δ is shown as a scatterplot of their NPVs. Every gray point corresponds to one phenotypic screen. Colored crosses highlight phenotypes suggestive of the genes’ shared function. (B) Similar to dap1Δ, yhr045wΔ is sensitive to DNA damaging agents hydroxyurea (HU) and methyl methanesulfonate (MMS). The sensitivity of the dap1Δ yhr045wΔ double mutant is identical to that of the two single mutants, suggesting that Dap1 and Yhr045w are epistatic to one another. (C) Dap1 is one of the five known physical interactors of Yhr045w. (D) The sensitivity of dap1Δ and yhr045wΔ to hydroxyurea and fluconazole is suppressed by the overexpression of ERG11. The sensitivity of dap1Δ and yhr045wΔ to hydroxyurea is suppressed by heme supplementation. (E) The similarity of the phenotypic profiles of ygl117wΔ and aro3Δ is shown as a scatterplot of their NPVs. Every gray point corresponds to one phenotypic screen. Colored crosses highlight phenotypes suggestive of the genes’ shared function. (F) The growth of ygl117w∆ is severely impaired in tryptophan-limited conditions (SC-Trp) relative to complete media (SC) but is restored in the absence of all three aromatic amino acids (SC-Trp-Tyr-Phe). YPD, yeast extract, peptone, and dextrose; WT, wild type; DMSO, dimethyl sulfoxide.