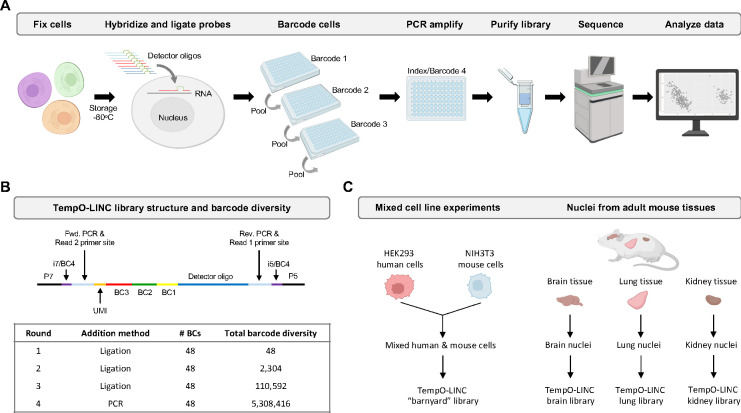
Figure 1. TempO-LINC workflow and study overview.
(A) The TempO-LINC workflow begins with fixation of cells or nuclei and subsequent storage at −80°C. Fixed cells are thawed and hybridized with Detector Oligo (DO) probe pools that are then ligated. Split pool barcoding with three rounds of ligation is then performed. Following ligation of barcodes, cells are pooled again and redistributed into a final 96-well plate where sequencing adaptors and index sequences are added during PCR amplification. Amplified libraries are then purified and sequenced. Data analysis begins with demultiplexing to associate gene expression profiles with cell-identifying barcodes. (B) Schematic of TempO-LINC 310 base pair library structure including the three barcodes that are ligated as well as the i5 and i7 index positions that serve as a fourth barcode to identify cells. The table shows total barcode diversity created through each round of barcoding as well as the molecular mechanism for barcoding. (C) Overview of TempO-LINC validation experiments performed for this study. Experiments were a combination of gene expression profiling on mixtures of multiple human and mouse cell lines as well as nuclei isolated from adult mouse brain, lung and kidney tissue.