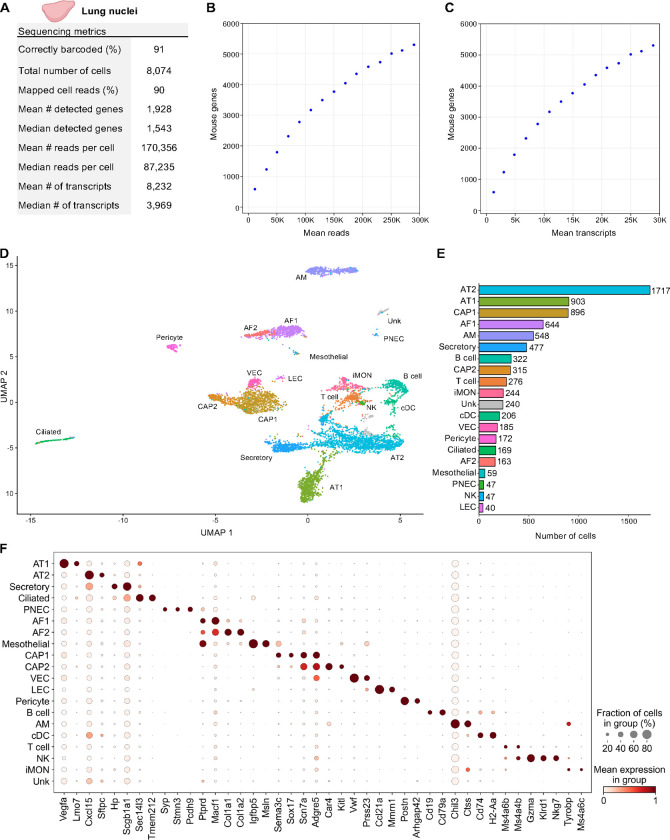
Figure 5. Single Nuclei Profiling of Adult Mouse Lung.
Nuclei from 8-week-old adult mouse lung tissue were barcoded with TempO-LINC. (A) Table of key performance metrics following sequencing and demultiplexing of lung nuclei libraries. (B,C) Sequencing saturation curves showing average number of genes detected per cell with increasing read or transcript depth. (D) 7,670 cells were plotted on a UMAP following unsupervised clustering. A total of 20 distinct populations were identified and annotated using data from the mouse LungMAP Consortium, available through the Lung Gene Expression Analysis (LGEA) web portal: alveolar type 1 (AT1), alveolar type 2 (AT2), capillary 1 (CAP1), capillary 2 (CAP2), venous endothelial cell (VEC), alveolar fibroblast 1 (AF1), alveolar fibroblast 2 (AF2), Secretory, Ciliated, Pericyte, alveolar macrophage (AM), B cell, T cell, inflammatory monocytes (iMON), classical dendritic cell (cDC), mesothelial (Mesothelial), natural killer (NK), lymphatic endothelial cell (LEC), pulmonary neuroendocrine cell (PNEC) and an unknown population (Unk). (E) Bar plot graph showing the number of cells within each population of clustered cells. (F) The expression level of 42 representative lung cell type biomarkers was examined for each of the 20 populations identified following unsupervised clustering.