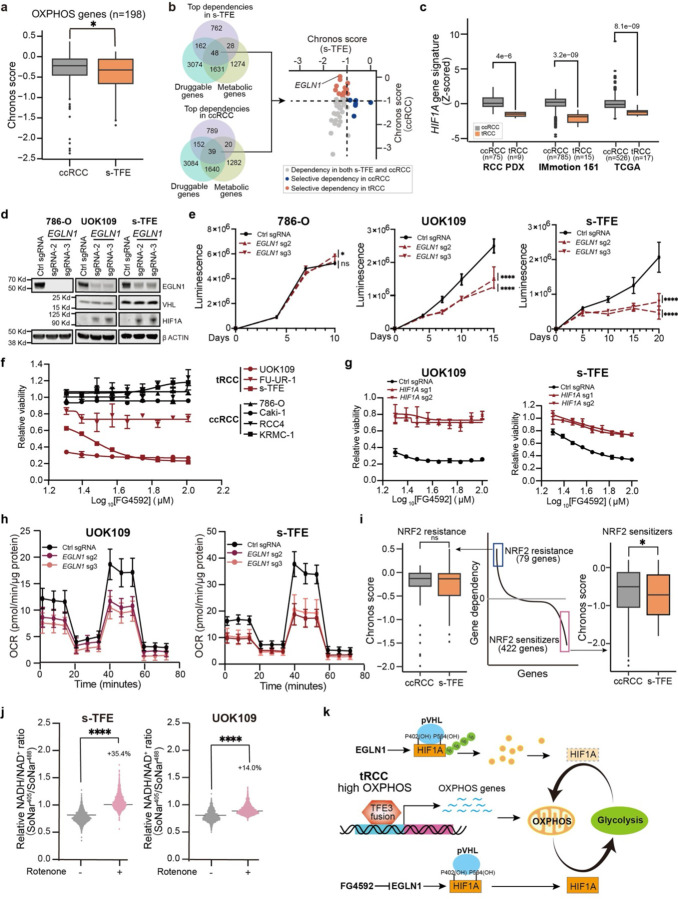
Fig. 3: Selective EGLN1 dependency in tRCC.
(a) Comparison of dependency scores for OXPHOS genes in ccRCC cell lines (averaged across 5 cell lines for each gene) vs. s-TFE tRCC cells.
(b) Gene dependencies identified via genome-scale CRISPR/Cas9 screening in s-TFE (tRCC) or ccRCC cells were overlapped with druggable and metabolic gene lists to nominate EGLN1 as a candidate selective dependency in tRCC cells.
(c) HIF1A gene signature scores in ccRCC or tRCC tumors from three independent studies as in Figure 1A.
(d) Western blot showing expression of EGLN1, VHL, and HIF1A after knockout of EGLN1 in a ccRCC cell line (786-O) or tRCC cell lines (UOK109 and s-TFE).
(e) Cell proliferation of tRCC cell lines (UOK109 and s-TFE) and ccRCC (786-O) after knockout of EGLN1. Data are shown as mean ± s.d. n=3 biological replicates.
(f) Relative cell viability (normalized to DMSO control) of tRCC cell lines or ccRCC lines after treatment with the EGLN inhibitor FG4592. Data are shown as mean ± s.d. n=3 biological replicates.
(g) Relative viability of HIF1A knockout tRCC cell lines (n=2, UOK109 and s-TFE) after treatment with FG4592. Data are shown as mean ± s.d. n=3 biological replicates.
(h) OCR after knockout of EGLN1 in UOK109 and s-TFE cell lines. Data shown as mean ± s.d, n=8–16 biological replicates.
(i) Genes whose knockout has been previously reported41 to confer sensitivity (422 genes) or resistance (79 genes) to NRF2 activation were compared for dependency in s-TFE cells vs. ccRCC cells (average of dependency score for 5 ccRCC cell lines, for each gene).
(j) Quantification of NADH to NAD+ ratio via SoNar assay following rotenone treatment in tRCC cell lines (UOK109 and s-TFE).
(k) Model: EGLN1 modulates OXPHOS and is a dependency in tRCC.
For panel (a), (c), (e), and (i-j), statistical significance was determined by Mann-Whitney U test. *p < 0.05, **p < 0.01, ***p < 0.001, **** p < 0.0001, n.s. not significant.