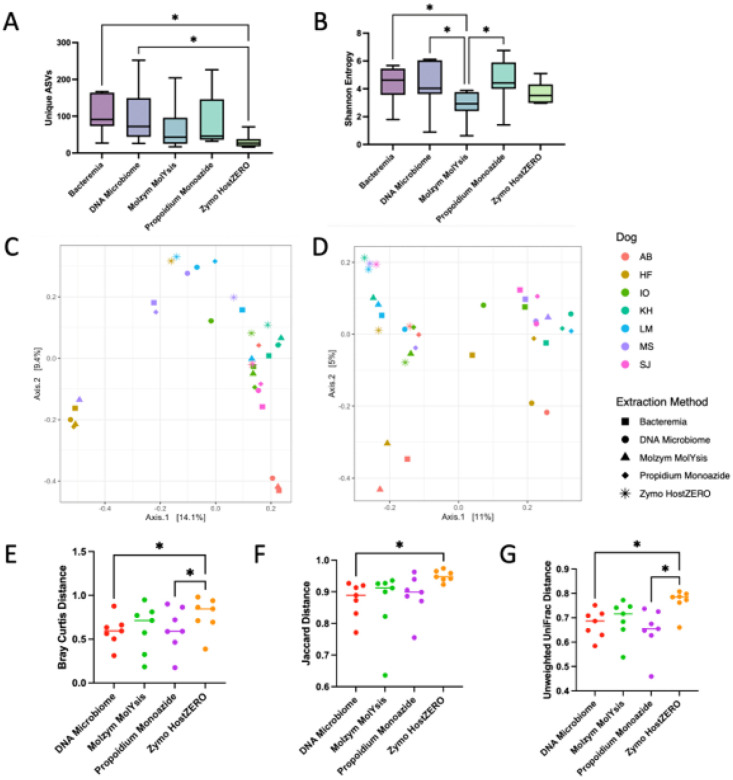
Figure 4. Microbial diversity and composition by extraction method (16S).
A) Microbial richness, or number of unique ASVs, and B) Microbial diversity (Shannon entropy) differed significantly by extraction method (Richness p=0.0018, Shannon p=0.0091, Friedman, multiple comparisons with FDR at 0.05, Table S8). Whiskers represent minimum, maximum, and median. *p<0.05. C) Microbial composition (Bray-Curtis) differed significantly by dog (PERMANOVA p=0.001), but not extraction method (PERMANOVA p=0.92) D) When microbial composition was weighted by phylogeny (Unweighted UniFrac), composition differed significantly by both extraction method (PERMANOVA p=0.002), and by dog (PERMANOVA p=0.001). E) Bray-Curtis, F) Jaccard, and G) Unweighted UniFrac distances from Bacteremia-extracted samples to samples of the same dog extracted via other extraction methods. E) Bray-Curtis F) Jaccard, and G) Unweighted UniFrac distances differed significantly with DNA Microbiome samples being the most similar (shortest distance) to Bacteremia-extracted samples (Friedman, Bray-Curtis p=0.034; Jaccard p=0.0342; Unweighted UniFrac p=0.0071). Pairwise comparison p-values are outlined in Table S9. *p<0.05. Bar represents median.