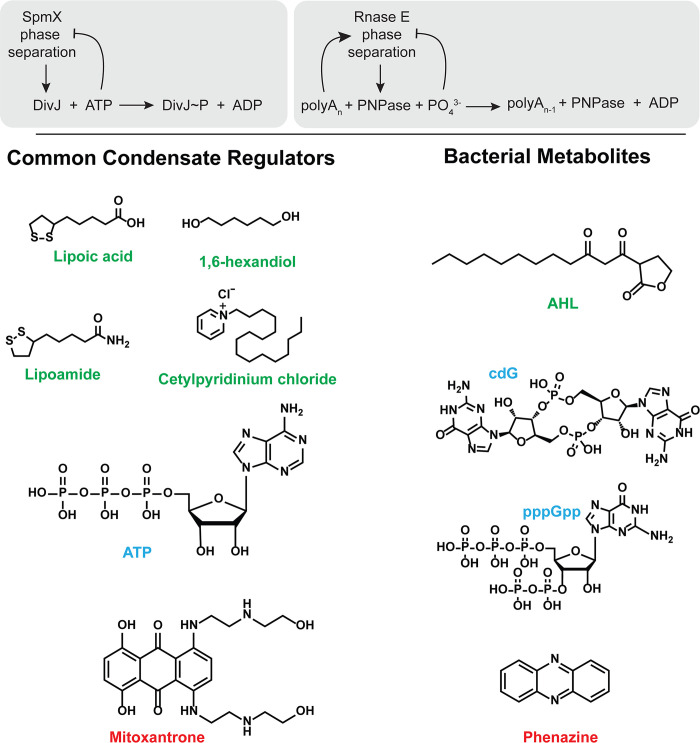
Fig 3. Small molecule-based regulation of condensate properties and function.
(Top) Salient examples of condensates from the aquatic bacterium Caulobacter crescentus that can be regulated by small molecule metabolites. SpmX is an IDP that forms polar condensates that localize the Histidine kinase DivJ. SpmX condensate formation is promoted under ATP depletion while dissolution is favored under high ATP concentrations. This ATP-dependent feedback is in turn exploited to regulate DivJ kinase activity in response to substrate availability. RNase E condensate regulates PNPase activity but is inhibited by phosphate, which is the substrate of this enzyme. RNase E phase separation is regulated by positive feedback from one substrate (polyA) and negative feedback from the other (phosphate). (Bottom) A repertoire of molecules that can tune condensate formation and disassembly. Lipoic acid, lipoamide, cetylpyridinium chloride, ATP, and hexanediol have been shown to dissolve biomolecular condensates. Other molecules with similar structural features such as antibiotics (Mitoxantrone), second messengers (cyclic di-GMP), and quorum-sensing ligands (AHLs) can be putative modulators of bacterial condensates.