FIGURE 1.
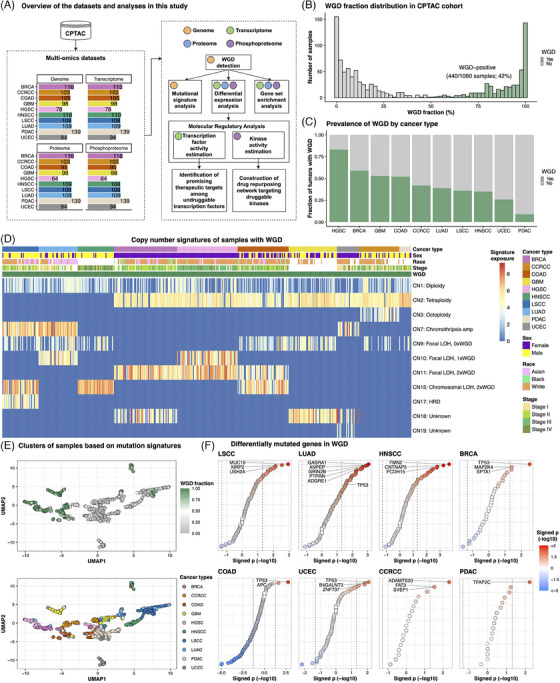
Copy number signatures associated with whole‐genome doubling (WGD). (A) Schematic representation of the multi‐omics datasets collected for each cancer type and the subsequent analysis pipeline. (B) Distribution of the WGD fraction within the Clinical Proteomic Tumour Analysis Consortium (CPTAC) cohort, displaying a bimodal pattern. (C) Prevalence of WGD by cancer type. (D) Heatmap based on copy number signature exposure values in samples with WGD. Copy number signatures were derived from the Catalogue of Somatic Mutations In Cancer (COSMIC). (E) Uniform manifold approximation and projection (UMAP) plot based on signature exposure values of single‐base‐substitution, double‐base‐substitution, indel and copy number alteration. Each dot indicates samples and is colored based on WGD fraction (top) and cancer types (bottom). (F) Dot plot depicting differentially mutated genes in WGD in each cancer type. High‐grade serous carcinoma (HGSC) and glioblastoma (GBM), which had no significantly enriched mutations in WGD, are not shown. The position of each dot along the x‐axis and the colour of the dots indicate signed p‐values obtained using Fisher's exact test. Only the top three significant genes were labelled in each cancer type, except for colon adenocarcinoma (COAD) and pancreatic ductal adenocarcinoma (PDAC), which had only two and one significant genes, respectively. In lung adenocarcinoma (LUAD), PTPRN, GRIN2B, ANPEP and ADGRE1 genes exhibited the same p‐values, and TP53 was additionally labelled. BRCA, breast cancer; CCRCC, clear cell renal cell carcinoma; HNSCC, head and neck squamous cell carcinoma; LSCC, lung squamous cell carcinoma; UCEC, uterine corpus endometrial carcinoma.
